Abstract
Regulated synthesis of reactive oxygen species (ROS) by membrane-bound fungal NADPH oxidases (Nox) plays a key role in fungal morphogenesis, growth, and development. Generation of reactive oxygen species (ROS) by the plant symbiotic fungus, Epichloë festucae, requires functional assembly of a multisubunit complex composed of NoxA, a regulatory component, NoxR, and the small GTPase RacA. However, the mechanism for assembly and activation of this complex at the plasma membrane is unknown. We found by yeast two-hybrid and coimmunoprecipitation assays that E. festucae NoxR interacts with homologs of the yeast polarity proteins, Bem1 and Cdc24, and that the Phox and Bem1 (PB1) protein domains found in these proteins are essential for these interactions. GFP fusions of BemA, Cdc24, and NoxR preferentially localized to actively growing hyphal tips and to septa. These proteins interact with each other in vivo at these same cellular sites as shown by bimolecular fluorescent complementation assays. The PB1 domain of NoxR is essential for localization to the hyphal tip. An E. festucae ΔbemA mutant was defective in hyphal morphogenesis and growth in culture and in planta. The changes in fungal growth in planta resulted in a defective symbiotic interaction phenotype. Our inability to isolate a Δcdc24 mutant suggests this gene is essential. These results demonstrate that BemA and Cdc24 play a critical role in localizing NoxR protein to sites of fungal hyphal morphogenesis and growth. Our findings identify a potential shared ancestral link between the protein machinery required for fungal polarity establishment and the Nox complex controlling cellular differentiation.
The NADPH oxidases (Nox) are a widely distributed family of eukaryotic proteins that transfer electrons across biological membranes to catalyze the reduction of molecular oxygen to superoxide (1–3). The multiple Nox isoforms found in eukaryotic cells control various physiological and cellular differentiation processes, including cell proliferation, apoptosis and hormone responses in animals (1, 2), and programmed cell death, hormone signaling and root hair tip growth in plants (4).
Fungi have three distinct subfamilies of NADPH oxidase (3, 5). NoxA has the core NADPH oxidase transmembrane and catalytic domains but no additional motifs, whereas NoxB has in addition, an N-terminal extension of ∼40 amino acids that is conserved among fungal species that have this isoform (3, 6). NoxC has a longer N-terminal extension, of 170–250 amino acids, which contains a putative calcium-binding EF-hand motif (3), similar to that found in human Nox5 and the plant Rboh enzymes (3, 5). In Aspergillus nidulans, Podospora anserina and Neurospora crassa, NoxA (Nox1) is required for the development of the sexual fruiting body, indicating that a common function of this isoform is regulation of multicellular development (7–10). NoxB (Nox2) is required for ascospore germination in P. anserina and N. crassa (7, 10). However, in Botrytis cinerea, both NoxA and NoxB are required for formation of the multicellular sclerotial sexual structure, but are dispensable for ascospore germination (11).
Fungal NADPH oxidases are also required for cellular growth and differentiation processes associated with plant host infection and colonization. In the symbiotic interaction between the endophytic fungus Epichloë festucae and perennial ryegrass, deletion of noxA, but not noxB, disrupted the highly regulated pattern of growth seen in WT associations (6). In the rice blast pathogen, Magnaporthe oryzae, disruption of either nox1 (noxA) or nox2 (noxB) resulted in loss of plant pathogenicity, due to an inability of the fungus to develop a penetration peg beneath the appressorium (12). Both NoxA and NoxB are important for pathogenicity of B. cinerea, but just NoxB is required for formation of the penetration structure (11).
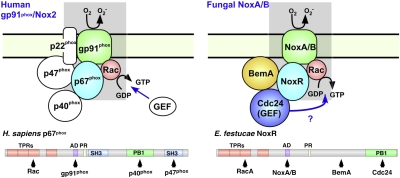
NoxR, a fungal homolog of the phagocytic p67phox Nox regulator, has been shown to regulate both NoxA and NoxB. In the symbiotic fungus E. festucae, a noxR mutant has a similar disrupted symbiotic interaction phenotype as ΔnoxA (6, 13). In B. cinerea and N. crassa, deletion of noxR (nor-1) resulted in a similar developmental phenotype to the noxA(nox1)/noxB(nox2) double mutant (7, 11). Although NoxA and NoxB have distinct cellular functions, it is not yet known how NoxR regulates these two different Nox isoforms. The N-terminal domain of NoxR is similar to p67phox, and includes four tetratricopeptide repeat (TPR) motifs required for Rac binding and a putative NADPH oxidase activation domain (3, 13) (Fig. 1A). In contrast, the C terminus of NoxR lacks the protein–protein interaction domains present in p67phox, including Src Homology 3 (SH3) and a conventional Phox and Bem1 (PB1), for interaction with p47phox and p40phox, respectively (1, 14). The absence of these domains in NoxR is consistent with the apparent absence of p47phox and p40phox homologs in fungal genome databases (3, 13). However, NoxR does possess a nonconventional PB1 domain in the C terminus of the protein, suggesting that fungi have distinct regulatory components that, upon activation, interact with NoxR to translocate this protein from the cytosol to the plasma membrane to assemble and activate the Nox enzyme complex (3, 13) (Fig. 1A).
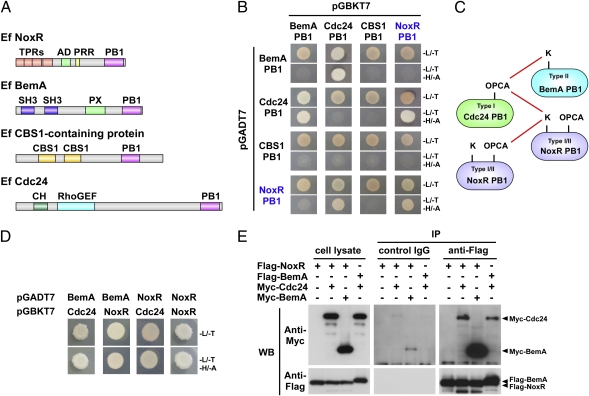
Fig. 1.
(A) Domain structure of E. festucae NoxR, BemA, Cdc24, and CBS1-containing protein. The tetratricopeptide repeat (TPRs), Nox activation (AD), proline-rich region (PRR), Src homology 3 (SH3), Phox and Bem1 (PB1), phox homology (PX), cystathionine beta-synthase 1 (CBS1), and calponin homology (CH) domains are indicated. (B) Yeast two-hybrid assay of the interactions between PB1 domains of E. festucae NoxR, BemA, Cdc24, and CBS1-containing protein. Yeast strain AH109 was transformed with prey and bait vectors, pGADT7 and pGBKT7, as indicated and plated on to SD medium lacking leucine and tryptophan (Upper, -Leu/-Trp) or lacking leucine, tryptophan, histidine, and adenine (Lower, -Leu/-Trp/-His/-Ade). Growth on the latter indicates an interaction between bait and prey. (C) Summary of interactions between PB1 domains based on results shown in B and Fig. S4A. (D) Yeast two-hybrid assay of interactions between full-length NoxR, BemA, and Cdc24. The plating conditions are the same as in B. (E) COS-7 cells were cotransfected with pEF-BOS-Flag-NoxR and pEF-BOS-Myc, or pEF-BOS-Myc-Cdc24 or pEF-BOS-Myc-BemA, or with pEF-BOS-Flag-BemA and pEF-BOS-Myc-Cdc24. Lysates of the transfected cells (Left) were analyzed by immunoprecipitation (IP) with control IgG (Center) or the anti-Flag monoclonal antibody (Right), followed by immunoblotting (WB) with the anti-Myc (Upper) or anti-Flag (Lower) monoclonal antibodies.
The objectives of this study were (i) to identify additional components of the fungal Nox enzyme complex by identifying proteins that interact with the PB1 domain of E. festucae NoxR, (ii) to investigate the subcellular localization of these regulators to identify where the Nox complex is assembled, and (iii) to determine whether these proteins, like NoxA and NoxR, have a role in maintaining the mutualistic symbiotic interaction between E. festucae and its host grass perennial ryegrass.
Results
Fungal Proteins That Interact with the C-Terminal PB1 Domain of NoxR.
The PB1 domain is a protein interaction domain conserved in eukaryotic cells (14). To identify candidate proteins that interact with NoxR via the PB1 domain, fungal genomes were interrogated by tBLASTN using the 27 different PB1 domain sequences from animal, yeast, and plant proteins compiled in the conserved protein domain database at NCBI. From this analysis, four PB1-containing proteins were consistently identified in fungal genome databases, NoxR itself, Cdc24, Bem1 (BemA) and a protein of unknown function containing tandem cystathionine beta-synthase 1 (CBS1) domains (Fig. 1A and Figs. S1–S3). During the course of this work, Kawahara and Lambeth (15) also identified these three proteins as candidate NoxR partners. Previous analysis indicated that the gene for NoxR is only present in fungal genomes that contain genes for NoxA/NoxB (3). However, genes for BemA, Cdc24, and the CBS1-containing protein are more widespread among fungal species, including those that lack a Nox complex, such as Saccharomyces cerevisiae and Ustilago maydis.
PB1 domains of NoxR, BemA, Cdc24 and CBS1-containing protein were cloned into yeast two-hybrid vectors pGADT7 and pGBKT7 and their interactions analyzed in yeast strain AH109. Interactions were detected between BemA-PB1 and Cdc24-PB1, Cdc24-PB1 and NoxR-PB1, and NoxR-PB1 and NoxR-PB1 (Fig. 1B). PB1 domains are classified into three types. Type I contains an acidic OPA, PC, and AID (OPCA) motif; type II contains an invariant Lys (K) residue on the first β-strand; and type I/II contains both motifs (14). Type I and type II PB1 domains interact with one another to form heterodimers, in which acidic residues on the OPCA motif form salt bridges with basic residues of the Type II PB1 domain. Analysis of amino acid sequence alignments of Cdc24, BemA and NoxR sequences identified PB1 domains of type I, type II, and type I/II, respectively (Figs. S1–S3). To verify the specificity of the interactions among BemA, Cdc24, and NoxR PB1 domains, site-directed mutants of each of these PB1 domains were tested for their ability to interact in a yeast two-hybrid assay (Fig. S4A). Taken together, these results confirm that BemA interacts with Cdc24, Cdc24 with NoxR and NoxR with NoxR via their respective PB1 domains (Fig. 1C).
The ability of full-length copies of Cdc24, BemA, and NoxR polypeptides to interact was also assessed. Interactions between full-length Cdc24 and BemA, Cdc24 and NoxR, and NoxR and NoxR were detected (Fig. 1D), results consistent with the interactions observed for their PB1 domains. Unexpectedly, an interaction between BemA and NoxR was detected even though the PB1 domains of these polypeptides did not interact (Fig. 1D). Yeast two-hybrid assays of various deletion derivatives of these proteins demonstrated that the PB1 domains of Cdc24 and BemA are also able to interact with regions of BemA and NoxR that lack PB1 domains, a result not totally without precedent (Fig. S4 B–D) (14). Coimmunoprecipitation assays confirmed interactions of Cdc24 with NoxR, Cdc24 with BemA, and BemA with NoxR (Fig. 1E).
Localization of NoxR, BemA, Cdc24, and RacA.
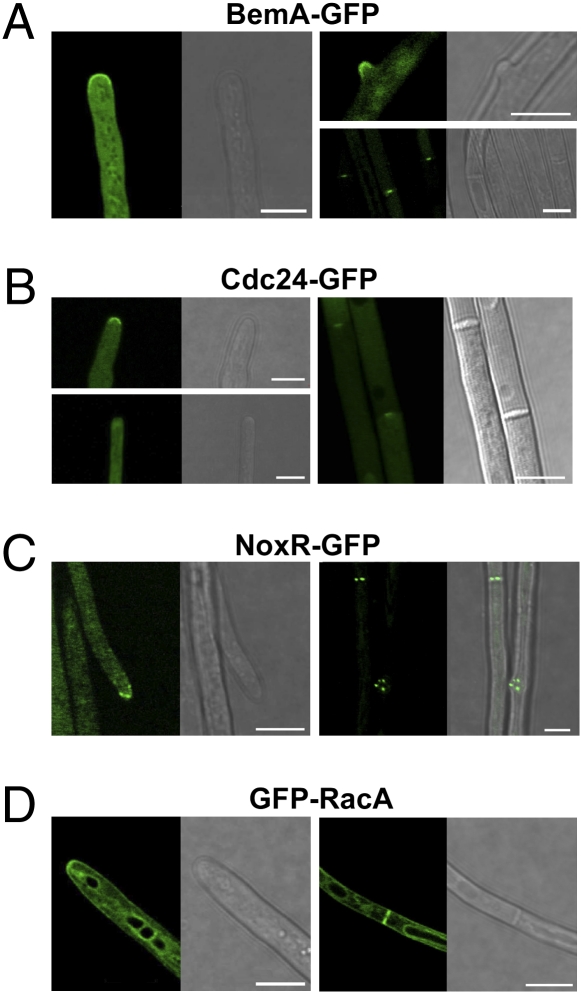
Previously, we showed that the E. festucae NoxA and the regulatory components NoxR and RacA are required to control hyphal tip growth and branching in culture and in the host plant (6, 13, 16). Nitroblue tetrazolium (NBT) staining of fungal cells in culture revealed that ROS are preferentially produced at hyphal tips (6), suggesting that assembly of the Nox complex occurs at hyphal tips and emerging branches. To test this hypothesis, GFP fusions of NoxR, RacA, BemA, and Cdc24 containing a 6-aa linker to allow appropriate folding of the two proteins, were expressed in E. festucae hyphae and localized within the cells by GFP fluorescence. The growth rate and hyphal morphology of all GFP transformants were the same as the WT. BemA-GFP consistently localized to the plasma membrane of hyphal tips, tips of emerging hyphal branches and at the center of hyphal septa (Fig. 2A and Fig. S5A). Transformants containing the Cdc24-GFP fusion consistently showed GFP fluorescence at the center of hyphal septa, but only infrequently localized to the plasma membrane of hyphal tips (Fig. 2B and Fig. S5B). NoxR-GFP, like BemA-GFP, consistently localized to hyphal tips and tips of emerging branches but the pattern of fluorescence was punctate, a pattern similar to that previously reported for polarity protein TeaR at hyphal tips of A. nidulans (Fig. 2C and Fig. S5C) (17). NoxR-GFP also localized to hyphal septa, again with a punctate pattern of distribution. GFP-RacA localized to the plasma membrane throughout the cell but was also localized to the plasma membrane of hyphal tips (Fig. 2D and Fig. S5D). Internal vacuolar membranes also fluoresced in the GFP-RacA transformants, an observation consistent with previous reports indicating that GTPases of the Rho family have been implicated in vacuole formation in S. cerevisiae and U. maydis (18, 19).
Fig. 2.
Subcellular localization of BemA-GFP, Cdc24-GFP, NoxR-GFP, and GFP-RacA in hyphae of E. festucae in axenic culture. BemA-GFP (A), Cdc24-GFP (B), NoxR-GFP (C), and GFP-RacA (D) proteins were expressed under the control of a TEF promoter in E. festucae. Localization of GFP-tagged proteins was observed for at least three individual transformants for each construct. Images shown are representative of the localization patterns observed for all constructs. (Bar, 5 μm.)
Nox Regulatory Components Interact in Vivo.
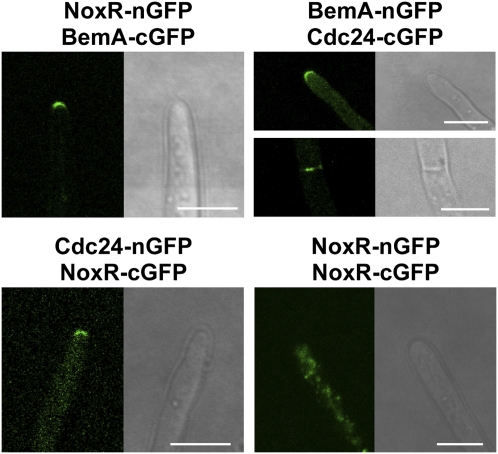
To test whether BemA, Cdc24, NoxR, and RacA interact in E. festucae, we generated fusions of these proteins with N- and C-terminal domains of GFP and assessed their interaction in hyphae using bimolecular fluorescence complementation (BiFC). Fluorescent arcs, indicative of interactions of NoxR with BemA, BemA with Cdc24, and Cdc24 with NoxR, were observed at hyphal tips (Fig. 3). Fluorescent complexes of Cdc24 with BemA were also observed at septa. In contrast, fluorescent complexes of NoxR homodimers were dispersed throughout the cytosol, rather than concentrated at the tip suggesting that heterodimer formation with other protein components is required for tip localization.
Fig. 3.
Bimolecular fluorescence complementation (BiFC) analysis of the interaction between BemA, Cdc24, and NoxR. BemA, Cdc24, or NoxR tagged with nGFP (2-174) or cGFP (175-239), were expressed in E. festucae under the control of the TEF promoter as indicated. Localization of fluorescence from GFP-tagged proteins was observed for at least three individual transformants for each construct. Images shown are representative of the localization patterns observed. (Bar, 5 μm.)
PB1 Domain Is Required for the Localization of NoxR at Hyphal Tips.
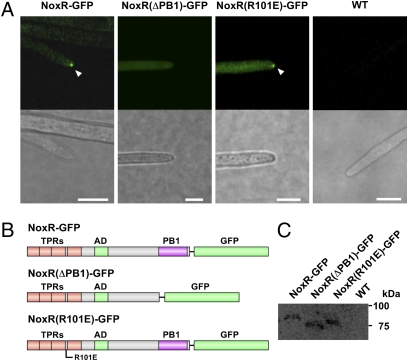
To identify the essential domain of NoxR required for localization of this protein to hyphal tips, mutant versions of NoxR-GFP were transformed into E. festucae. Previously, we showed that substitution of a conserved arginine (R101), between the third and fourth TPR domains of NoxR, for a glutamic acid, compromised interaction of NoxR with RacA (13). Fluorescence imaging showed that NoxR(R101E)-GFP still localized at the hyphal tip, demonstrating that binding of RacA is not essential for NoxR to localize at the hyphal tip. In contrast, NoxR lacking the PB1 domain and tagged with GFP, NoxR(ΔPB1)-GFP, showed no GFP fluorescence at the hyphal tip (Fig. 4 A and B). Western analysis showed that lack of localization was not due to a lack of stable expression of the truncated protein (Fig. 4C). These results indicate that the PB1 domain is essential for localization of NoxR to the hyphal tip.
Fig. 4.
PB1 domain is essential for localization of NoxR at hyphal tips. (A) NoxR with a single amino acid substitution in the RacA binding site (R101E) and NoxR lacking the PB1 domain (ΔPB1), were tagged with GFP and expressed in E. festucae under the control of the TEF promoter. Localization of GFP-tagged proteins was observed for at least three individual transformants for each construct. Images shown are representative of the localization patterns observed for all constructs. (B) Domain structure of E. festucae GFP-tagged NoxR, NoxR (ΔPB1), or NoxR (R101E). (C) Expression of GFP-tagged NoxR or NoxR variants in transformants. Protein samples were extracted from hyphae of WT (WT) or transformants expressing NoxR-GFP, NoxR (ΔPB1)-GFP, or NoxR(R101E)-GFP. Expressed GFP-tagged proteins (50 μg in each lane) were detected by immunoblot analysis with anti-GFP antibody. (Bar, 5 μm.)
Deletion of E. festucae bemA Disrupts Normal Symbiotic Interaction with Host.
To investigate the biological role of BemA and Cdc24 in axenic culture and plant symbiosis, replacement constructs for bemA (pPN152) and cdc24 (pPN153) were prepared and linear fragments of these plasmids recombined into the genome of E. festucae Fl1 (Fig. S6). Hygromycin resistant transformants of bemA were screened by PCR, and one putative replacement, bemA-139 (ΔbemA), was identified from 200 transformants analyzed. Southern blot analysis of genomic digests of the ΔbemA mutant probed with pPN153, combined with additional PCR analysis, confirmed a replacement event at the bemA locus (Fig. S6). PCR screening of 212 hygromycin resistant cdc24 transformants failed to identify a replacement mutant. Given cdc24 mutants of U. maydis and Candida albicans are unviable (20, 21), it is likely that deletion of this gene in E. festucae is also lethal.
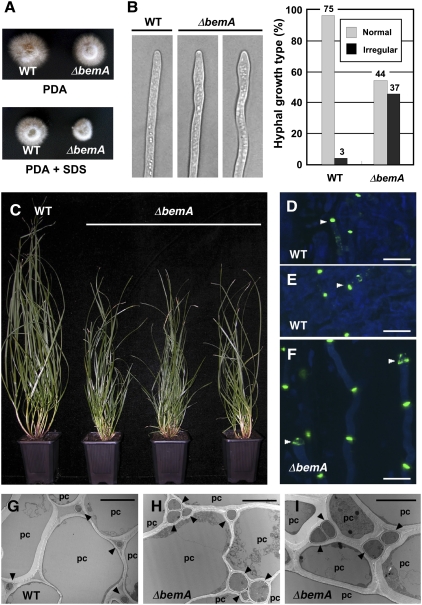
Although there were no obvious differences in colony morphology between the WT strain and the ΔbemA mutant grown on PD agar, in the presence of 0.01% SDS the ΔbemA mutant showed reduced radial growth compared with the WT, indicating a defect in cell wall integrity (Fig. 5A). Light microscopy analysis revealed that the subapical region of the hyphal tips of the ΔbemA mutant, were frequently swollen (Fig. 5B) and the hyphae variable in width. This phenotype was particularly pronounced when the ΔbemA mutant was grown on SDS-containing media. Transformation of the ΔbemA mutant with pNPP13, containing the full-length bemA gene, restored the WT culture phenotype (Figs. S6 and S7). No differences were detected between the WT and ΔbemA in hyphal ROS production in culture using a sensitive chemiluminescence method, probably due to the very low level of ROS production by this fungus.
Fig. 5.
Axenic culture and in planta phenotype of E. festucae bemA mutant. (A) Colony morphology of WT Fl1 (WT) and ΔbemA mutant on PD agar (Upper) and PD agar with 0.01% SDS (Lower). (B) DIC images of WT Fl1 (WT) and ΔbemA mutant hyphal tips after growth on PD agar for 7 d and percentages of normal and irregularly shaped hyphal tips of both strains. Number of hyphae counted is given above each column. (C) Phenotype of perennial ryegrass plants infected with E. festucae WT Fl1 (WT) and ΔbemA mutant strains. Photograph was taken 12 wk after inoculation. (D–F) Confocal micrographs of aniline blue/WGA-AF488–stained hyphae growing in leaf sheath of 14-wk-old perennial ryegrass plants infected with WT Fl1 (WT; D and E) and ΔbemA mutant (F) strains. (Bar, 50 μm.) (G–I) Transmission electron micrographs of cross-sections of E. festucae Fl1 WT (WT; G) and ΔbemA mutant strain (H and I) hyphae (arrowheads) in the intercellular space of perennial ryegrass. pc, Plant cell. Photograph was taken 12 wk after inoculation. (Bar, 2 μm.)
To determine the symbiotic interaction phenotype of the ΔbemA mutant, seedlings of perennial ryegrass were infected with mutant and WT strains. In contrast to the normal growth of host plants infected with WT strain, perennial ryegrass infected with ΔbemA had a mildly stunted phenotype (Fig. 5C), in contrast to the severe stunting observed for the ΔnoxA mutant (6). Aniline blue/WGA-AF488 staining of infected plant tissues revealed that some hyphal tips of the ΔbemA mutant were swollen in planta (Figs. 5 D–F). Transmission electron microscopy analysis of hyphae in the intercellular space of the outer leaf sheath showed that ΔbemA mutant hyphae were both larger in diameter and more abundant than in WT associations (Fig. 5 G–I and Fig. S7 D and E). Plants infected with the ΔbemA/bemA complemented strains displayed a WT symbiotic interaction phenotype (Fig. S7C).
To examine the effect of the bemA mutation on NoxR localization, hyphae containing stable NoxR-GFP fluorescence complexes at the tip, as imaged by confocal microscopy, were counted in the WT and bemA mutant backgrounds. This analysis showed there was a significant reduction in formation of stable NoxR complex formation at the hyphal tip (Fig. S8). These results, taken together with the yeast two-hybrid analysis (Fig. S4), suggest that Cdc24 is likely to be the essential component for hyphal tip localization of NoxR but that BemA plays an important auxiliary role.
Discussion
We have shown here that E. festucae NoxR, an essential regulatory component of the filamentous fungal NADPH oxidase (NoxA/NoxB) complex (7, 11, 13), physically interacts with homologs of the yeast polarity establishment proteins Bem1 and Cdc24 (22). A survey of available fungal genomes revealed that genes for NoxA/NoxB, NoxR, RacA, BemA, and Cdc24 are widely distributed within the fungal kingdom (3, 15), suggesting that this group of proteins comprises an ancestral enzyme complex for ROS production. In those fungal lineages that lack genes for NoxA/NoxB, NoxR, and RacA (e.g., Saccharomycotina), homologs of Bem1 and Cdc24 appear to have been retained as part of the core machinery for polarity establishment. We have shown here that the PB1 domains of NoxR, Cdc24 and BemA are essential for the interaction of these proteins both in vitro and in vivo. The PB1 domain, originally defined because of its presence in proteins of the mammalian phagocytic oxidase (Phox) and yeast polarity establishment (Bem-related proteins) complex, is a protein interaction module conserved in animals, fungi, protists, and plants (14). Our findings identify a potential shared ancestral link between the protein machinery required for fungal polarity establishment and the Nox complex controlling cellular differentiation.
Activation of Nox2 (gp91phox) in mammalian phagocytes requires translocation of p67phox from the cytosol to the plasma membrane (1, 2). The recruitment of p67phox, together with the other cytosolic components p40phox and p47phox, involves a complex series of protein–protein interactions (2). Like p67phox, fungal NoxR has no predicted transmembrane or membrane-associated domains, so requires a scaffold protein(s) functionally equivalent to the mammalian Nox organizers, p47phox and p40phox, for its recruitment to the plasma membrane to activate NoxA/NoxB. The BemA protein is a strong candidate to fulfill this function given the demonstration here that it physically interacts with NoxR, and contains protein interaction domains, SH3, PB1, and PX, similar to those found in p47phox and p40phox, which would facilitate interactions with other proteins and the plasma membrane (23).
The varied physical interactions detected for NoxR, BemA, and Cdc24 suggest that fungi, like mammals, have developed a complex regulatory system for Nox activation in response to external stimuli (2). The presence of a typeI/II PB1 domain in NoxR allows formation of both homo- and heterodimers. The self-interaction of NoxR provides strong evidence for homodimerization in vivo, which appears to be predominantly cytosolic as demonstrated here by BiFC. In metabolically active cells, NoxR preferentially localizes as patches to hyphal tips and septa. Deletion of the NoxR PB1 domain compromises tip localization. Given that Cdc24 physically interacts with NoxR and that the PB1 domain of NoxR is not essential for interaction of NoxR with BemA, heterodimerization of Cdc24 and NoxR is probably the critical step required for recruitment of NoxR to the hyphal tip. The preferential localization of NoxR, Cdc24 and BemA to septa may indicate that these proteins provide a mark for subapical polarity, mark branch points, or store these proteins at these sites to be available for polarized growth of branch points. Cdc24 is a guanine–nucleotide exchange factor (GEF) that catalyzes the exchange of small GTPase-bound GDP by GTP. Cdc42, the yeast homolog of RacA, is a substrate of Cdc24 (24). In U. maydis, Don1 was identified as the GEF for Cdc42, and Cdc24 is proposed as the GEF for RacA to control the processes of cell septation and polarized growth, respectively (18, 25, 26). We previously showed that interaction of the GTP bound form of RacA with NoxR is essential for activation of the fungal Nox (13, 16). Thus, Cdc24 is likely to be the GEF for activation of RacA in the fungal Nox complex. Given that binding of RacA to NoxR is not necessary for NoxR localization at the hyphal tip, the GEF activity of Cdc24 is not directly required to control NoxR localization. Because BemA also preferentially localizes to the hyphal tip and is able to interact with both Cdc24 and NoxR, this adaptor protein is likely to be the scaffold that facilitates recruitment of NoxR-Cdc24 to the apical tip. This hypothesis is supported by the requirement for BemA for the preferential and stable localization of NoxR to the hyphal tip. Anchoring of the BemA protein in the plasma membrane is likely to be mediated by the PX domain, a domain found in p40phox and p47phox that is known to bind specific phospholipids to facilitate assembly of the mammalian Nox complex (27, 28). Sterol-rich lipid rafts, rich in sphingolipids, have been shown to concentrate at the growing tips of hyphae from C. albicans and A. nidulans (29, 30) and to be required for polarized localization of the cell end marker proteins, TeaA and TeaR, in A. nidulans (31). These may be the sites for PX domain anchoring of BemA to assemble the Nox complex at the hyphal tip. However, given that the ΔbemA mutant maintains relatively normal radial growth in E. festucae and A. nidulans (32, 33), additional proteins are likely to be involved, or are able to substitute for BemA, in the recruitment of Cdc24 and the other components required for fungal Nox complex assembly and activation.
Attempts to isolate a deletion mutant of cdc24 in E. festucae were unsuccessful, presumably because Cdc24 is essential for growth as reported for cdc24 replacement mutants in S. cerevisiae, U. maydis and Ashbya gossypii (22, 34, 35). In contrast, the E. festucae ΔbemA mutant had relatively normal hyphal development with the exception of a slight loss in cell wall integrity, as detected by sensitivity to SDS, and swollen hyphal tips. An alteration in hyphal tip shape was also observed for the A. nidulans ΔbemA mutant (32, 33); a phenotype proposed to be caused by destabilization of the polarisome (32), a complex required for controlling tip shape (36). Bem1 is required to maintain the localization of polarity regulators at the polarisome in S. cerevisiae (37). Its absence would lead to a breakdown of the positive feedback loop required to reinforce stabilization of this complex. The symbiotic interaction phenotype of the E. festucae ΔbemA mutant is also mild compared with the severe interaction phenotype previously reported for the ΔnoxA, ΔnoxR, and ΔracA mutants (6, 13, 16). Although multiple, enlarged hyphae are observed in the intercellular spaces of the leaves, the plants do not undergo premature senescence as observed for the ΔnoxA, ΔnoxR, and ΔracA mutants. The relatively mild phenotype of the E. festucae bemA mutant is consistent with the less severe clinical phenotype of chronic granulomatous disease (CGD) observed for human mutations in the Nox organizer subunit p47phox compared with gp91phox (Nox2) (38).
In U. maydis, polar localization of Cdc24 is primarily dependent on Bem1, but if overexpressed in a bem1-defective background, it still localizes to the hyphal tip, suggesting there is redundancy in the tip localization machinery (20). Targeting of Cdc24 to the hyphal tip, where it physically interacts and colocalizes with Bem1, requires the cyclin-dependent kinase Cdk5 (20). Many proteins are required for polarity establishment in fungi, including SpaA, BudA, KipA, TeaA, and TeaR in A. nidulans (17, 31). Mutation of any of these genes causes a partial loss of polarity, resulting in a zig-zag or curved pattern of hyphal growth but retention of filamentous growth (17). These results suggest that there are overlapping mechanisms for establishment and maintenance of the polarized growth pattern characteristic of filamentous fungi.
In summary, we have shown that BemA and Cdc24, two well-characterized regulators of polarity establishment in yeast, are, in addition, important components of the filamentous fungal NADPH oxidase enzyme complex (Fig. 6). We propose that Cdc24 is the GEF for activation of RacA and that BemA has an analogous role to the mammalian p40phox and p47phox “organizer” proteins to recruit and assemble NoxR and RacA at the plasma membrane to activate NoxA/NoxB (Fig. 6). Identifying the mechanisms responsible for activating and mobilizing the cytoplasmic components of the Nox complex to the plasma membrane, and how the ROS generated by this complex, control fungal polarized growth, and cellular differentiations will be important areas for further investigation.
Fig. 6.
NADPH oxidase complexes in fungal and human phagocytic cells. (A) Human gp91phox and the regulatory components for ROS production in phagocytic cells. (B) Model for regulation of NoxA/B catalyzed ROS production by recruitment of RacA and NoxR to the fungal plasma membrane. NoxR interacts with RacA via the TPR domains and with Cdc24 via the PB1 domain. NoxR also interacts with the PB1 domain of BemA. Domain structures of fungal NoxR and human p67phox are shown below diagrams of fungal and human Nox complexes, respectively. Adapted from model previously proposed by Takemoto et al. (3).
Materials and Methods
Biological material (Table S1), primers (Table S2), plasmids (Tables S3 and S4), molecular biology methods, PCR conditions, preparation of constructs, transformation, yeast two-hybrid, DNA sequencing, bioinformatics, immunoprecipitation and microscopy are described in detail in SI Materials and Methods.
Binding Assays.
Yeast two-hybrid assays using pGADT7 or pGBKT7 (Clontech) based constructs were performed according to the manufacturer's instructions (MATCHMAKER Two-Hybrid System3). Immunoprecipitation analysis was performed with COS-7 cells transfected with pEF-BOS-FLAG or pEF-BOS-Myc based vectors by method adapted from Izaki et al. (39).
Microscopy.
Images of GFP-tagged proteins in E. festucae hyphae were captured using a confocal laser scanning microscope FV1000-D (Olympus) using the default settings for enhanced GFP. Transmission electron microscopy, light microscopy for toluidine blue staining, NBT staining, and aniline blue staining were performed as previously described (6).
Acknowledgments
We thank Douglas Hopcroft, Carla Eaton and Emma Brasell (Massey University), Michael Christensen and Wayne Simpson (AgResearch) for technical assistance. We thank Carla Eaton for comments on the manuscript, Murray Cox for assistance with statistical analysis and Steve Harris for comments on septal localization of proteins. The E. festucae E2368 genome sequence was made available by Christopher Schardl through Grants EF-0523661 from the US National Science Foundation (to Christopher Schardl, Mark Farman, and Bruce Roe) and 2005-35319-16141 from the US Department of Agriculture National Research (to Christopher Schardl). This research was supported by the Tertiary Education Commission to the Bio-Protection Research Centre, the Royal Society of New Zealand Marsden Fund Grant 07MAU046, and a Grant-in-Aid for Young Scientists 20780032 (to D.T.) from the Japan Society for the Promotion of Science.
Footnotes
The authors declare no conflict of interest.
Data deposition: The sequences reported in this paper have been deposited in the DDBJ/EMBL/GenBank databases (accession nos. AB524338 and AB524339).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017309108/-/DCSupplemental.
References
- 1.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nat Rev Immunol. 2004;4:181–189. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 2.Sumimoto H. Structure, regulation and evolution of Nox-family NADPH oxidases that produce reactive oxygen species. FEBS J. 2008;275:3249–3277. doi: 10.1111/j.1742-4658.2008.06488.x. [DOI] [PubMed] [Google Scholar]
- 3.Takemoto D, Tanaka A, Scott B. NADPH oxidases in fungi: Diverse roles of reactive oxygen species in fungal cellular differentiation. Fungal Genet Biol. 2007;44:1065–1076. doi: 10.1016/j.fgb.2007.04.011. [DOI] [PubMed] [Google Scholar]
- 4.Torres MA, Dangl JL. Functions of the respiratory burst oxidase in biotic interactions, abiotic stress and development. Curr Opin Plant Biol. 2005;8:397–403. doi: 10.1016/j.pbi.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 5.Aguirre J, Ríos-Momberg M, Hewitt D, Hansberg W. Reactive oxygen species and development in microbial eukaryotes. Trends Microbiol. 2005;13:111–118. doi: 10.1016/j.tim.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 6.Tanaka A, Christensen MJ, Takemoto D, Park P, Scott B. Reactive oxygen species play a role in regulating a fungus-perennial ryegrass mutualistic interaction. Plant Cell. 2006;18:1052–1066. doi: 10.1105/tpc.105.039263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cano-Domínguez N, Alvarez-Delfín K, Hansberg W, Aguirre J. NADPH oxidases NOX-1 and NOX-2 require the regulatory subunit NOR-1 to control cell differentiation and growth in Neurospora crassa. Eukaryot Cell. 2008;7:1352–1361. doi: 10.1128/EC.00137-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lalucque H, Silar P. NADPH oxidase: An enzyme for multicellularity? Trends Microbiol. 2003;11:9–12. doi: 10.1016/s0966-842x(02)00007-0. [DOI] [PubMed] [Google Scholar]
- 9.Lara-Ortíz T, Riveros-Rosas H, Aguirre J. Reactive oxygen species generated by microbial NADPH oxidase NoxA regulate sexual development in Aspergillus nidulans. Mol Microbiol. 2003;50:1241–1255. doi: 10.1046/j.1365-2958.2003.03800.x. [DOI] [PubMed] [Google Scholar]
- 10.Malagnac F, Lalucque H, Lepère G, Silar P. Two NADPH oxidase isoforms are required for sexual reproduction and ascospore germination in the filamentous fungus Podospora anserina. Fungal Genet Biol. 2004;41:982–997. doi: 10.1016/j.fgb.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 11.Segmüller N, et al. NADPH oxidases are involved in differentiation and pathogenicity in Botrytis cinerea. Mol Plant Microbe Interact. 2008;21:808–819. doi: 10.1094/MPMI-21-6-0808. [DOI] [PubMed] [Google Scholar]
- 12.Egan MJ, Wang Z-Y, Jones MA, Smirnoff N, Talbot NJ. Generation of reactive oxygen species by fungal NADPH oxidases is required for rice blast disease. Proc Natl Acad Sci USA. 2007;104:11772–11777. doi: 10.1073/pnas.0700574104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Takemoto D, Tanaka A, Scott B. A p67Phox-like regulator is recruited to control hyphal branching in a fungal-grass mutualistic symbiosis. Plant Cell. 2006;18:2807–2821. doi: 10.1105/tpc.106.046169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sumimoto H, Kamakura S, Ito T. Structure and function of the PB1 domain, a protein interaction module conserved in animals, fungi, amoebas, and plants. Sci STKE. 2007 doi: 10.1126/stke.4012007re6. 10.1126/stke.4012007re6. [DOI] [PubMed] [Google Scholar]
- 15.Kawahara T, Lambeth JD. Molecular evolution of Phox-related regulatory subunits for NADPH oxidase enzymes. BMC Evol Biol. 2007 doi: 10.1186/1471-2148-7-178. 10.1186/1471-2148-7-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanaka A, Takemoto D, Hyon GS, Park P, Scott B. NoxA activation by the small GTPase RacA is required to maintain a mutualistic symbiotic association between Epichloë festucae and perennial ryegrass. Mol Microbiol. 2008;68:1165–1178. doi: 10.1111/j.1365-2958.2008.06217.x. [DOI] [PubMed] [Google Scholar]
- 17.Fischer R, Zekert N, Takeshita N. Polarized growth in fungi—interplay between the cytoskeleton, positional markers and membrane domains. Mol Microbiol. 2008;68:813–826. doi: 10.1111/j.1365-2958.2008.06193.x. [DOI] [PubMed] [Google Scholar]
- 18.Mahlert M, Leveleki L, Hlubek A, Sandrock B, Bölker M. Rac1 and Cdc42 regulate hyphal growth and cytokinesis in the dimorphic fungus Ustilago maydis. Mol Microbiol. 2006;59:567–578. doi: 10.1111/j.1365-2958.2005.04952.x. [DOI] [PubMed] [Google Scholar]
- 19.Müller O, Johnson DI, Mayer A. Cdc42p functions at the docking stage of yeast vacuole membrane fusion. EMBO J. 2001;20:5657–5665. doi: 10.1093/emboj/20.20.5657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Tabarés I, Pérez-Martín J. Cdk5 kinase regulates the association between adaptor protein Bem1 and GEF Cdc24 in the fungus Ustilago maydis. J Cell Sci. 2008;121:2824–2832. doi: 10.1242/jcs.026286. [DOI] [PubMed] [Google Scholar]
- 21.Bassilana M, Blyth J, Arkowitz RA. Cdc24, the GDP-GTP exchange factor for Cdc42, is required for invasive hyphal growth of Candida albicans. Eukaryot Cell. 2003;2:9–18. doi: 10.1128/EC.2.1.9-18.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Madden K, Snyder M. Cell polarity and morphogenesis in budding yeast. Annu Rev Microbiol. 1998;52:687–744. doi: 10.1146/annurev.micro.52.1.687. [DOI] [PubMed] [Google Scholar]
- 23.Marcotte EM, et al. Detecting protein function and protein-protein interactions from genome sequences. Science. 1999;285:751–753. doi: 10.1126/science.285.5428.751. [DOI] [PubMed] [Google Scholar]
- 24.Zheng Y, Cerione R, Bender A. Control of the yeast bud-site assembly GTPase Cdc42. Catalysis of guanine nucleotide exchange by Cdc24 and stimulation of GTPase activity by Bem3. J Biol Chem. 1994;269:2369–2372. [PubMed] [Google Scholar]
- 25.Hlubek A, Schink KO, Mahlert M, Sandrock B, Bölker M. Selective activation by the guanine nucleotide exchange factor Don1 is a main determinant of Cdc42 signalling specificity in Ustilago maydis. Mol Microbiol. 2008;68:615–623. doi: 10.1111/j.1365-2958.2008.06177.x. [DOI] [PubMed] [Google Scholar]
- 26.Weinzierl G, et al. Regulation of cell separation in the dimorphic fungus Ustilago maydis. Mol Microbiol. 2002;45:219–231. doi: 10.1046/j.1365-2958.2002.03010.x. [DOI] [PubMed] [Google Scholar]
- 27.Ago T, et al. Phosphorylation of p47phox directs phox homology domain from SH3 domain toward phosphoinositides, leading to phagocyte NADPH oxidase activation. Proc Natl Acad Sci USA. 2003;100:4474–4479. doi: 10.1073/pnas.0735712100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kanai F, et al. The PX domains of p47phox and p40phox bind to lipid products of PI(3)K. Nat Cell Biol. 2001;3:675–678. doi: 10.1038/35083070. [DOI] [PubMed] [Google Scholar]
- 29.Li S, Du L, Yuen G, Harris SD. Distinct ceramide synthases regulate polarized growth in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2006;17:1218–1227. doi: 10.1091/mbc.E05-06-0533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Martin SW, Konopka JB. Lipid raft polarization contributes to hyphal growth in Candida albicans. Eukaryot Cell. 2004;3:675–684. doi: 10.1128/EC.3.3.675-684.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Takeshita N, Higashitsuji Y, Konzack S, Fischer R. Apical sterol-rich membranes are essential for localizing cell end markers that determine growth directionality in the filamentous fungus Aspergillus nidulans. Mol Biol Cell. 2008;19:339–351. doi: 10.1091/mbc.E07-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Leeder AC, Turner G. Characterisation of Aspergillus nidulans polarisome component BemA. Fungal Genet Biol. 2008;45:897–911. doi: 10.1016/j.fgb.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 33.Semighini CP, Harris SD. Regulation of apical dominance in Aspergillus nidulans hyphae by reactive oxygen species. Genetics. 2008;179:1919–1932. doi: 10.1534/genetics.108.089318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Castillo-Lluva S, Alvarez-Tabarés I, Weber I, Steinberg G, Pérez-Martín J. Sustained cell polarity and virulence in the phytopathogenic fungus Ustilago maydis depends on an essential cyclin-dependent kinase from the Cdk5/Pho85 family. J Cell Sci. 2007;120:1584–1595. doi: 10.1242/jcs.005314. [DOI] [PubMed] [Google Scholar]
- 35.Wendland J, Philippsen P. Cell polarity and hyphal morphogenesis are controlled by multiple rho-protein modules in the filamentous ascomycete Ashbya gossypii. Genetics. 2001;157:601–610. doi: 10.1093/genetics/157.2.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Taheri-Talesh N, et al. The tip growth apparatus of Aspergillus nidulans. Mol Biol Cell. 2008;19:1439–1449. doi: 10.1091/mbc.E07-05-0464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Butty AC, et al. A positive feedback loop stabilizes the guanine-nucleotide exchange factor Cdc24 at sites of polarization. EMBO J. 2002;21:1565–1576. doi: 10.1093/emboj/21.7.1565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Berg JM, et al. Chronic granulomatous disease: The European experience. PLoS ONE. 2009 doi: 10.1371/journal.pone.0005234. 10.1371/journal.pone.0005234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Izaki T, Kamakura S, Kohjima M, Sumimoto H. Two forms of human Inscuteable-related protein that links Par3 to the Pins homologues LGN and AGS3. Biochem Biophys Res Commun. 2006;341:1001–1006. doi: 10.1016/j.bbrc.2006.01.050. [DOI] [PubMed] [Google Scholar]