Abstract
Phosphorothioate (PT) modification of DNA, with sulfur replacing a nonbridging phosphate oxygen, was recently discovered as a product of the dnd genes found in bacteria and archaea. Given our limited understanding of the biological function of PT modifications, including sequence context, genomic frequencies, and relationships to the diversity of dnd gene clusters, we undertook a quantitative study of PT modifications in prokaryotic genomes using a liquid chromatography-coupled tandem quadrupole mass spectrometry approach. The results revealed a diversity of unique PT sequence contexts and three discrete genomic frequencies in a wide range of bacteria. Metagenomic analyses of PT modifications revealed unique ecological distributions, and a phylogenetic comparison of dnd genes and PT sequence contexts strongly supports the horizontal transfer of dnd genes. These results are consistent with the involvement of PT modifications in a type of restriction-modification system with wide distribution in prokaryotes.
Keywords: DNA modification, bioanalytical chemistry, sulfur
Phosphorothioate (PT) modification of DNA, in which sulfur replaces a nonbridging phosphate oxygen, was originally developed as an artificial means to stabilize oligodeoxynucleotides against nuclease degradation (1). However, we recently discovered that the dnd gene products incorporate sulfur into the DNA backbone as a PT in a sequence- and stereo-specific manner (2). Beginning with the original observation in Streptomyces lividans 1326 that the five-gene dnd cluster (dndA–E) caused DNA degradation during electrophoresis (3), the presence of dnd genes has been established in dozens of different bacteria and archaea (4). An emerging picture of Dnd protein function reveals that DndA acts as a cysteine desulfurase and assembles DndC as a 4Fe-4S cluster protein (5). DndC possesses ATP pyrophosphatase activity and is predicted to have PAPS reductase activity, whereas DndB has homology to a group of transcriptional regulators (4, 6). A DndD homologue in Pseudomonas fluorescens Pf0-1, SpfD, has ATPase activity possibly related to DNA structure alteration or nicking during PT incorporation (7).
This progress in defining the biochemistry of PT modifications belies a lack of understanding of the biological function of PT modifications, such as the variety of sequence contexts, the distribution of modifications across prokaryotic genomes, and the relationship of PT sequence contexts to the diversity of known dnd gene clusters (4). We have approached this problem with a highly quantitative study of PT modifications in prokaryotic genomes using a liquid chromatography-coupled tandem quadrupole mass spectrometry (LC-MS/MS) approach. The results reveal a diversity of quantized PT sequence contexts consistent with a role for PT modifications as part of a restriction-modification system.
Results and Discussion
Development of a Sensitive Method to Quantify PT Modifications in Bacterial Genomes.
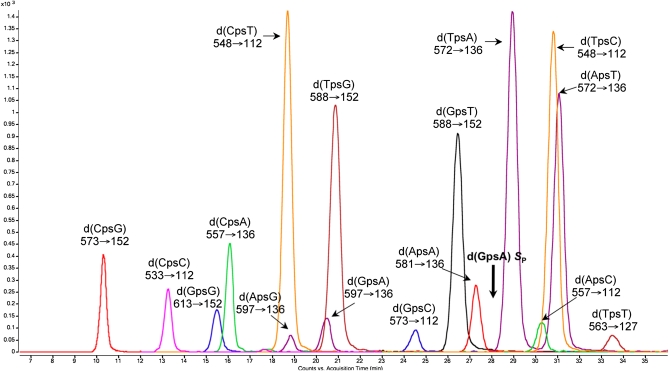
We approached the problem of defining the biological function of PT modifications by quantifying them and defining their sequence context. Specifically, we developed a highly quantitative electrospray ionization LC-MS/MS technique that identifies the two-nucleotide sequence context of PT modification. By using synthetic dinucleotides containing PT in the RP configuration (the only stereochemistry observed to date), the HPLC retention times, collision-induced dissociation molecular transitions, and limits of detection were optimized for all 16 possible dinucleotide sequence contexts for PT (Fig. 1 and Table S1). We also optimized conditions for nuclease P1, which is inhibited by PT in the RP configuration, and alkaline phosphatase hydrolysis of genomic DNA to PT-containing dinucleotides and canonical nucleosides (Fig. S1), with canonical nucleosides eluting well before the PT-modified dinucleotides. Quantification was achieved by using the SP stereoisomer of d(GPSA) as an internal standard with multiple reaction monitoring mode of the mass spectrometer (Fig. 1). We were thus able to detect PT modifications at levels as low as 1 per 106 nt for d(TPST) to 2 per 108 nt for d(CPST) in 20 μg of genomic DNA (Table S1). This quantitative bioanalytical approach is a rigorous means to screen prokaryotic genomes for the sequence context and quantity of PT modifications, with immediate implications for the understanding of biological function.
Fig. 1.
Analysis of PT-linked dinucleotides by LC-MS/MS in multiple reaction monitoring mode. All of the 16 possible PT-linked dinucleotides in RP configuration were resolved by reversed-phase HPLC followed by MS/MS detection by using the ion transitions labeled under each dinucleotide. Bold arrow indicates d(GPSA) SP internal standard.
Widespread Distribution and Diverse Sequence Contexts for PT Modifications in Bacteria and Archaea.
The LC-MS/MS method was first applied to define the PT sequence contexts and quantify PT modifications in bacteria known to harbor dnd gene clusters. These taxonomically unrelated bacteria, including Salmonella enterica serovar Cerro 87, Escherichia coli B7A, P. fluorescens Pf0-1, S. lividans 1326, Geobacter uraniumreducens Rf4, Hahella chejuensis KCTC2396, Bermanella marisrubri RED65, and Shewanella pealeana ATCC700345, represent genera of variable origins and diverse habitats, such as soil-dwelling and marine microbes, aerobic and anaerobic microbes, nonpathogenic saprophytes, and human pathogens. One immediate discovery was the presence of a PT sequence context, d(GPST), in bacteria originally observed to possess d(GPSA) (Table 1) (2). In the cases of S. enterica serovar Cerro 87 and E. coli B7A, d(GPST) and d(GPSA) occurred at the same level of approximately 4 PT per 104 nt (Table 1). However, this pairing of PT sequence contexts was not universal, with marine bacteria B. marisrubri RED65 and H. chejuensis KCTC2396 possessing d(GPSA) accompanied by only barely detectable d(GPST) and S. pealeana ATCC700345 possessing a ratio of d(GPSA) to d(GPST) of approximately 2:1 (Table 1). Apart from the combination of d(GPST) and d(GPSA), both d(GPST) and d(GPSG) were simultaneously present in G. uraniumreducens Rf4 and S. lividans 1326, but at levels that differed by two orders of magnitude (Table 1). Distinct from the others, P. fluorescens Pf0-1 possesses only a single PT context, d(GPSG), at a level close to that in S. lividans 1326 and G. uraniumreducens Rf4. These results in bacteria known to possess dnd genes revealed the potential for a wide range of sequence contexts for PT modifications, as might be expected for a restriction-modification system.
Table 1.
PT modifications of DNA in bacteria
| PT modifications per 106 nt |
|||||||
| Bacterium | d(GPSA) | d(GPST) | d(GPSG) | d(CPSA) | d(APSA) | d(TPSA) | Total PT |
| E. coli B7A | 370 ± 11 | 398 ± 17 | — | — | — | — | 768 ± 27 |
| S. enterica 87 | 362 ± 9 | 370 ± 11 | — | — | — | — | 732 ± 20 |
| DH10B (pJTU1980) | 529 ± 48 | 543 ± 62 | — | 2 ± 0 | 3 ± 0 | 2 ± 0 | 1,078 ± 109 |
| DH10B (pJTU1238) | 717 ± 52 | 774 ± 53 | — | 4 ± 0 | 6 ± 1 | 5 ± 0 | 1,505 ± 103 |
| P. fluorescens Pf0-1 | — | — | 451 ± 9 | — | — | — | 451 ± 9 |
| S. lividans 1326 | — | 2 ± 0 | 471 ± 39 | — | — | — | 474 ± 39 |
| G. uraniumreducens Rf4 | — | 3 ± 0 | 517 ± 14 | — | — | — | 520 ± 13 |
| B. marisrubri RED65 | 438 ± 23 | 3 ± 0 | — | — | — | — | 440 ± 23 |
| S. pealeana ATCC700345 | 316 ± 9 | 172 ± 2 | — | — | — | — | 489 ± 11 |
| H. chejuensis KCTC2396 | 286 ± 9 | * | — | — | — | — | 286 ± 9 |
Values represent mean ± SD for three analyses of 20 μg of bacterial DNA; dash indicates that the dinucleotide was not detected.
*Signal for the dinucleotide was detected but below the limit of quantification.
Further insights into PT function were gained from surveys of bacteria not previously known to possess dnd genes. To this end, we interrogated 63 Vibrio strains derived from an approximate 1,000-strain library of ecologically differentiated coastal Vibrionaceae with defined seasonal and habitat preferences (8). Seven isolates were found to possess PT modifications, with levels of four to six per 104 nt in d(GPSA)/d(GPST) and d(GPSG) in three strains (1F267, ZS139, 1F230). Strikingly, four other isolates possessed 10-fold higher levels of PT (2–3 per 103 nt) in a new sequence context, d(CPSC) (Table 2). Partial genome sequence information for these isolates revealed dnd gene homologues in three of the strains (1F267, ZS139, and 1F230; Materials and Methods). These results both expand the repertoire of sequence contexts and broaden the range of PT levels in bacterial genomes to cover three orders of magnitude.
Table 2.
PT modifications in DNA from Vibrio isolates
| Modifications per 106 nt |
Modifications per 107 nt |
|||||
| Vibrio isolates | d(GPSA) | d(GPST) | d(GPSG) | d(CPSC) | d(APSC) | d(TPSC) |
| 1F267 | 289 ± 21 | 287 ± 27 | — | — | — | — |
| ZS139 | — | 19 ± 1 | 499 ± 18 | — | — | — |
| 1F230 | — | 3 ± 0 | 397 ± 5 | — | — | — |
| 1C-10 | — | — | — | 3,107 ± 71 | 19 ± 2 | 12 ± 0 |
| ZF264 | — | — | — | 2,269 ± 18 | 8 ± 2 | 2 ± 0 |
| ZF29 | — | — | — | 2,240 ± 57 | 10 ± 1 | 4 ± 0 |
| FF75 | — | — | — | 2,624 ± 22 | 11 ± 2 | 4 ± 1 |
Values represent mean ± SD for three analyses of 20 μg of bacterial DNA; dash indicates that the dinucleotide was not detected.
PT Modifications Are Quantized in Three Discrete Frequencies.
Analysis of the quantitative data revealed that the levels of PT modifications were quantized into three distinct levels: two to three per 103 nt, three to eight per 104 nt, and one to six per 106 nt (Tables 1 and 2). Along with defined sequence contexts, the first two frequency ranges are consistent with a restriction-modification system (9). The highest frequency of two to three PT modifications per 103 nt (one PT modification in 333–500 nt) was observed in Vibrio species as d(CPSC) (Table 2), which is consistent with a 4-nt consensus sequence, such as GGCPSC or CPSCGG, with a statistical frequency of once every 256 nt. Analysis of available partial genome sequence data for these strains (Table S2) reveals that the CCGG motif occurs every approximately 905 ± 130 nt and the GGCC motif every approximately 518 ± 54 nt (averages for four species), whereas the CC dinucleotide motif occurs every 24 nt. With allowance for the A/T richness of the Vibrio genomes (Table S2), the GGCC and CCGG motifs are thus reasonable candidates for palindromic PT consensus sequences.
The other PT frequency consistent with a restriction-modification function is three to eight PT per 104 nt (Tables 1 and 2), which is equivalent to one PT in 1,250 to 3,333 nt or a 5–6-nt consensus sequence (∼1 modification every 1,024–4,096 nt). This frequency was observed for the d(GPSG) motif in S. lividans (Table 1) (2), in which a loose consensus sequence of 5′-cGGCCgccg-3′ (GGCC strictly conserved) was determined based on cloning of dnd phenotype break sites (10, 11). Analysis of the limited genome sequence data available for S. lividans 1326 reveals that the strictly conserved 4 nt GPSGCC core sequence occurs at too high a frequency (one site per ∼110 nt; Table S3) to serve as the consensus sequence for a PT-based restriction-modification system. Such is also the case for P. fluorescens Pf0-1 and G. uraniumreducens Rf4, in which GGCC occurs, on average, every 114 nt and 177 nt, respectively (Table S3). Therefore, a 6-nt consensus, such as CGPSGCCG that occurs at a frequency of 6 to 11 sites per 104 nt in S. lividans 1326, P. fluorescens Pf0-1, and G. uraniumreducens Rf4 (Table S3), is more consistent with the observed levels of PT modifications (three to eight per 104 nt).
A similar situation holds for E. coli B7A and S. enterica 87, in which we observed a 1:1 ratio of d(GPSA) and d(GPST) with each PT-containing site occurring every 2,500 to 2,800 nt on average (Table 1). This frequency is too low for a 4-nt consensus sequence such as GPSAAC/GPSTTC, which occurs in the related E. coli DH10B once in every 258 nt (Table S4), but it is consistent with a 1- to 2-nt extension of this core sequence. The situation with B. marisrubri RED65 and H. chejuensis KCTC2396, in which d(GPSA) was observed as the predominant dinucleotide context at frequencies consistent with a 5- to 6-nt consensus (Table 1), suggests a palindromic core sequence such as GPSATC.
One consequence of the high sensitivity of the LC-MS/MS method is that we were able to detect PT modifications at levels well below those expected for a restriction-modification system. As shown in Tables 1 and 2, we observed two to three PT modifications per 106 nt in the d(GPST) motif in S. lividans 1326 and G. uraniumreducens Rf4 for which d(GPSG) was the high frequency modification site, whereas d(CPSA), d(TPSA), and d(APSA) occurred at low levels (two to six per 106 nt) in E. coli expressing the S. enterica dnd genes (Table 1) and d(APSC) and d(TPSC) were minor sites observed in Vibrio isolates in Table 2.
There are two explanations for the low PT levels: a function for PT other than restriction-modification, as is the case for DNA methylation in many prokaryotes (9, 12); and biochemical nonspecificity for the Dnd protein responsible for target selection. The latter would be similar to a restriction enzyme cleaving at a thermodynamically or kinetically disfavored site (i.e., secondary cleavage sites), or cleaving with altered sequence specificity as a result of salt- or pH-induced alterations in protein–DNA interactions (i.e., “star activity”) (9). As one test of the latter hypothesis, we compared the levels of d(GPST) and d(GPSA) in E. coli DH10B harboring low and high copy number plasmids containing the S. enterica 87 dnd gene cluster, pJTU1980 and pJTU1238, respectively. The high copy number vector produced a 16-fold increase in dndC transcription (Table S5). As shown in Table 1, the increased expression of dnd genes caused PT modifications in d(GPST) and d(GPSA) to increase 1.5- and 2-fold, respectively, in comparison with S. enterica 87 (Table 1). This is consistent with relaxation of a strict modification consensus sequence or an increase in the proportion of a consensus sequence population that becomes modified with a PT. Support for the former model comes from the appearance of three low-frequency PT modifications in d(CPSA), d(TPSA), and d(APSA) in direct proportion with dnd gene expression (Table 1). The results support the hypothesis that low-frequency PT modifications result from a degree of relaxed DNA target recognition by Dnd proteins.
Phylogenetic Analysis of dnd Genes and PT Sequence Contexts Is Consistent with Horizontal Gene Transfer.
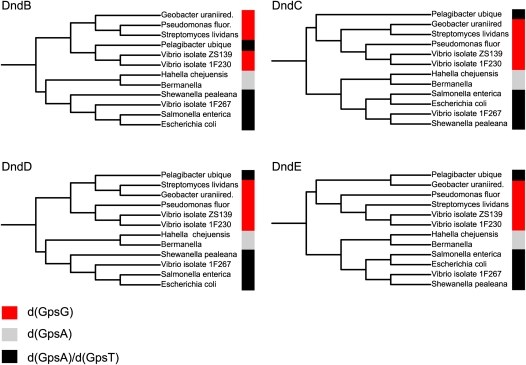
In addition to quantized PT levels, support for the involvement of PT modifications in a restriction-modification system comes from an apparent association of PT sequence contexts with phylogenetic relationships drawn from Dnd protein sequences but not species phylogeny. There is strong evidence for the distribution of classical methylation-based restriction-modification systems (13) and for the distribution of dnd gene clusters (14) by horizontal gene transfer and mobile genetic elements, as opposed to the vertical gene transfer associated with species phylogeny. As shown in Fig. 2, there is a strong correlation between the Dnd protein sequence phylogeny and the distribution of PT sequence modifications, with the exception of outlier Candidatus Pelagibacter ubique. The fact that the Dnd phylogenies do not follow their corresponding species tree suggests that PT sequence context is dependent on Dnd protein sequence and not on the phylogenetic descent of the strains. This is most clearly seen in the three Vibrio isolates (ZS139, 1F230, and 1F267), which are phylogenetically incoherent in all four Dnd proteins. The phylogenetic differentiation of the Vibrio isolates also suggests horizontal gene transfer of the whole dnd cluster. The fact that this split is matched by a corresponding switch in PT sequence context is a strong indication of the dependency between dnd genes and PT contexts.
Fig. 2.
Correlation between PT sequence contexts and Dnd protein sequences. Phylogenetic analysis reveals a correlation between PT sequence context and four of the Dnd protein sequences, and supports horizontal rather than vertical gene transfer for dnd genes. Dnd protein sequences for 12 strains analyzed here were retrieved from the NCBI Protein database and aligned using Muscle (35) and phylogenetic trees were computed with phyML (36), with phylogenies displayed using iTol (37). The PT sequence context is color-coded as noted in the key.
Insights into Ecological Distributions of PT Modifications and dnd Genes by Metagenomic Analysis of Ocean Genomes.
The widespread distribution of dnd genes and PT modifications is further illustrated by a metagenomic analysis of ocean bacteria (15). To initiate these studies, we identified a dnd gene cluster homologue in C. Pelagibacter ubique strain HTCC1002, one of the smallest known free-living bacteria and the first cultured member of the alphaproteobacterial SAR11 clade. SAR11 is a ubiquitous group of marine bacteria that can account for as much as 35% of bacterioplankton populations in the ocean surface (16). The SAR11 dnd gene cluster was located in a hypervariable region of the genome and was not found in closely related strain HTCC1062, which is consistent with our observation of PT only in strain HTCC1002 as d(GPSA) and d(GPST) RP modifications.
Having established a PT modification in strain HTCC1002, we used tBLASTn with the HTCC1002 dndBCDE genes as queries to detect the distribution of dnd genes in oceanic metagenomes. We observed significant hits (E<10−20) in several oceanic metagenomes (Table S6): 136 reads contained dndB, 123 reads contained dndC, 80 reads contained dndD, and nine reads contained dndE. In particular, one of the most significant hits was detected in the metagenome of Sargasso Sea, a low-nutrient, low-productivity, subtropical ocean gyre. DNA samples collected from depths to 200 m in the Sargasso Sea revealed natural DNA PT contexts of d(CPSC) and d(GPSA) in all samples, with d(GPST) and d(GPSG) occurring mainly at lower depths (Table 3). A similar analysis of DNA samples collected from 5 m, 20 m, and 40 m in the highly productive, temperate Oregon coastal waters revealed the presence of four major PT contexts: d(CPSC), d(GPSA), d(GPST), and d(GPSG). Interestingly, d(CPSC) and d(GPSA) were detected throughout the water columns off the Oregon coast (5–40 m) and the Sargasso Sea (0–200 m), whereas d(GPSG) was detected only in deeper zones of the water column in both locations. Dramatic transitions in the composition of microbial communities as a function of depth have been documented in previous studies (17). These results suggest that d(GPSG) is associated with microbial taxa that occupy the dark mesopelagic ocean region beneath the euphotic zone, which may have implications for horizontal gene transfer among microbes occupying the various ocean communities.
Table 3.
PT detection in seawater at varying depths
| DNA sample location/depth | d(CPSC) | d(GPSA) | d(GPSG) | d(GPST) |
| Oregon coast | ||||
| 5 m | + | + | — | — |
| 20 m | + | + | + | — |
| 40 m | + | + | + | + |
| Sargasso Sea | ||||
| 0 m | + | + | — | — |
| 80 m | + | + | — | — |
| 200 m | + | + | + | — |
Plus sign denotes detectable levels of PT in 2 μg of DNA; dash indicates that the PT-containing dinucleotide was not detectable. Oregon samples were collected from station SH50 (44° 15′ N, 124° 10′ W) on April 2, 2007; Sargasso Sea samples were collected at the BATS station on August 7, 2001 (15).
In summary, these results provide insights into the function of PT modifications in bacterial genomes. Our LC-MS/MS approach to studying the only defined chemical modification of the DNA backbone, with potential application to the study of putative arsenic-modified microbial nucleic acids (18), provides a rich source of information that complements genetic and molecular approaches to defining biological function. The data reveal quantized levels of PT, which suggest involvement of PT in a new restriction-modification system. This is consistent with our recent observation that a dptF-H cluster adjacent to dnd genes in S. enterica serovar Cerro 87 restricts the uptake of plasmids without PT (19) and with studies showing inhibition of methylation-based restriction enzymes by PT modifications (20, 21). The observations of widespread ecological distribution and strong phylogenetic relationships of dnd genes and PT modifications demonstrate the importance of PT modifications in prokaryotic physiology.
Materials and Methods
Materials, Bacterial Strains, and Plasmids.
Enantiomerically pure, PT-containing dinucleotides in Rp or Sp configuration were obtained from IBA BioTagnology. The bacteria strains harboring natural sets of dnd clusters and DNA samples were gifts from different laboratories and institutions as noted in the Acknowledgments. Plasmid pJTU1238 was a derivative of high copy plasmid pBluescript II SK+ containing dnd gene cluster from S. enterica 87 (2). A KpnI-XbaI fragment containing the dnd gene cluster from pJTU1238 was cloned into pIJ2925 to yield pJTU1976, from which the KpnI-BglII fragment harboring the dnd genes was inserted into the KpnI- and BamHI-treated low-copy plasmid pACYC184 to yield pJTU1980. We have also characterized the PT modifications in a set of well characterized Vibrionaceae strains (8) for which partial genome sequence information has been obtained.
E. coli B7A is an enterotoxigenic strain isolated from an American soldier suffering from diarrhea during the Vietnam War (22) and represents a frequent etiologic agent for short-incubation travelers diarrhea and endemic infantile diarrhea (23). S. enterica 87 was isolated from a commercial egg-producing farm (24). Marine bacteria H. chejuensis KCTC2396, B. marisrubri RED65, and S. pealeana ATCC700345 were isolated from marine sediment of Cheju Island in Korea, surface seawater from Gulf of Eilat in Red Sea, and the accessory nidamental gland of the squid Loligo pealei, respectively (25–27). G. uraniumreducens Rf4 was isolated from sediments of the Old Rifle uranium bioremediation field site and is an anaerobic organism capable of U(IV) reduction, showing potential for removing toxic uranium from contaminated groundwater (28). S. lividans 1326 is a soil actinomycete (29).
SAR11 Clade Growth and DNA Isolation.
SAR11 strains HTCC1002 and HTCC1062 were grown at 16 °C in filtered, autoclaved seawater amended with carbon, nitrogen, and phosphorus as described previously (30). Water was also amended with vitamins, 50 nM 3-dimethylsulphoniopropionate, and 50 nM glycine betaine. Cell growth was monitored using a Guava EasyCyte flow cytometer and cells were harvested in early stationary phase. DNA was extracted using the sucrose lysis method followed by phenol extraction and ethanol precipitation (31). DNA was further purified by treatment with RNase and extraction by using a Qiagen DNeasy Blood and Tissue Kit. Strain identities were verified by amplification and sequencing of the oxidoreductase gene as described previously (32).
Controlled Enzymatic Digestion of PT Modified DNA.
The first step in the development of the LC-MS/MS technique to quantify PT modifications involved nuclease P1-mediated hydrolysis of PT-containing DNA to a limit digest of nucleosides and PT-bridged dinucleotides, as observed in earlier studies (2). Optimal parameters for nuclease P1 hydrolysis were defined by using 20 μg of E. coli B7A DNA treated with varying quantities of nuclease P1 (0.25–8 U) in 30 mM sodium acetate, pH 5.3, 0.5 mM ZnCl2 in a 100-μL volume at 50 °C for incubations lasting 15 min to 8 h. Subsequent dephosphorylation was carried out by addition of 10 μL of 1 M Tris-Cl, pH 8.0, and 10 U of alkaline phosphatase at 37 °C for another 2 h. The enzymes were subsequently removed by ultrafiltration (YM-10 column; Microcon) followed by addition of 50 pmol of d(GPSA) SP as reference. The quantity of d(GPSA) in the naturally occurring RP configuration was monitored by LC-MS/MS to measure the hydrolytic efficiency, with 2 U of nuclease P1 and a 2-h incubation found to provide complete release of PT-containing dinucleotides.
LC-MS/MS Analysis of PT-Containing Dinucleotides.
The next step in developing an analytical method for PT modifications involved definition of chromatographic and MS parameters for resolving and quantifying the PT-containing dinucleotides. To this end, we first defined the HPLC retention times for the set of all possible 16 PT-linked dinucleotides by using a Thermo Hypersil GOLD aQ column (150 × 2.1 mm, 3 μm) with elution conducted at 35 °C and a flow rate of 0.3 mL/min, with a gradient of 97% buffer A (0.1% acetic acid in water) and 3% buffer B (0.1% acetic acid in acetonitrile) for 5 min, followed by 3% to 6% buffer B over a period of 30 min and 6% to 98% buffer B over a period of 1 min. Retention times are noted with other parameters later, with canonical nucleosides eluting as follows: dC, 2.2 min; dA, 3.8 min; dG, 3.9 min; and dT, 5.0 min; the d(GPSA) SP reference eluted at 28.1 min. The HPLC column was coupled to an Agilent 6410 Triple Quad LC/MS mass spectrometer with an electrospray ionization source in positive mode with the following parameters: gas flow, 10 L/min; nebulizer pressure, 25 psi; drying gas temperature, 300 °C; and capillary voltage, 3,100 V. Multiple reaction monitoring mode was used for detection of product ions derived from the precursor ions, with all instrument parameters optimized for maximal sensitivity (retention time in min, precursor ion m/z, product ion m/z, fragmentor voltage, collision energy): d(CPSG), 10.3, 573, 152, 123 V, 25 V; d(CPSC), 13.3, 533, 112, 123 V, 25 V; d(GPSG), 15.5, 613, 152, 123 V, 29 V; d(CPSA), 16.1, 557, 136, 126 V, 29 V; d(CPST), 18.7, 548, 112, 110 V, 13 V; d(APSG), 18.8, 597, 136, 120 V, 40 V; d(GPSA), 20.5, 597, 136, 120 V, 40 V; d(TPSG), 20.9, 588, 152, 117 V, 17 V; d(GPSC), 24.5, 573, 112, 129 V, 25 V; d(GPST), 26.5, 588, 152, 110 V, 17 V; d(APSA), 27.3, 581, 136, 117 V, 33 V; d(TPSA), 28.9, 572, 136, 125 V, 20 V; d(APSC), 30.3, 557, 112, 117 V, 25 V; d(TPSC), 30.8, 548, 112, 117 V, 13 V; d(APST), 31.1, 572, 136, 125 V, 20 V; and d(TPST), 33.5, 563, 127, 110 V, 37 V.
RNA Preparation and Real-Time Quantitative PCR.
To assess the expression of dnd genes, total RNA was isolated by using a Qiagen RNeasy Protect Bacteria Mini Kit and 15 ng of RNA was used as template for real-time PCR performed with the Power SYBR Green RNA-to-CT 1-Step Kit (Applied Biosystems) and an Applied Biosystems 7900HT Fast real-time PCR system. To measure the transcription of dnd cluster in DH10B(pJTU1238) and DH10B(pJTU1980), primers were designed within dndC gene. The housekeeping gene gapA, which codes for d-GAPDH, was used as reference. Primers 5′-ATTGTTGTCCTCGGTTCACG-3′ and 5′-GGCGGTATTTGAGCCAGTAG-3′ were used to amplify dndC gene; primers 5′- CCGTATCGGTCGCATTGTT-3′ and 5′-CTTCGTCCCATTTCAGGTT-3′ were used to amplify gapA gene. RT-PCR data analysis was performed according to the comparative threshold cycle method, also known as 2–ΔΔCT.
Partial Sequencing of Vibrio Genomes.
Each genome was sequenced using the Illumina platform with an independent lane of sequence for each, yielding approximately 12 million 76-bp single-end reads per genome. We removed read fragments containing Ns and trimmed poor-quality read termini with Euler qualityTrimmer (using default parameters) (33). The Velvet software package was used for de novo assembly of reads into contigs (34). Parameters were optimized to yield the most contiguous (i.e., highest N50) assembly, independently for each strain. N50 values ranged from 3 to 200 kb, with an average strain reaching an N50 of 40 kb and N80 of 20 kb.
Analysis of Bacterial Genomes.
Bacterial strains were analyzed for sequence motifs by using the DiProGB genome browser with sequences obtained from the National Center for Biotechnology Information (NCBI) Genome database. Complete or partial genome sequences were uploaded into the browser and the number of sequence motifs determined for both genomic DNA strands (Table S2). Dnd protein sequences for 12 strains analyzed here were retrieved from the NCBI Protein database and aligned by using Muscle (35) and phylogenetic trees were computed with phyML (36), with phylogenies displayed using iTol (37).
Acknowledgments
The authors thank Dr. Evgenya Shelobolina for providing G. uraniumreducens Rf4, Dr. Jian-Shen Zhao for S. pealeana ATCC700345, Dr. Hong Kum Lee for H. chejuensis KCTC2396, Dr. Eduardo Robleto for P. fluorescens Pf0-1, Dr. Toshiyuki Murase for S. enterica serovar Cerro 87, Joshua Kitner for culturing the SAR11 cells, Anthony Bertagnolli and the crew of the R.V. Elakha for collecting and processing Oregon Coast DNA samples, Rachel Parsons and the crew of the R.V. Weatherbird II for collecting and processing BATS samples, and Dr. H. James Tripp for assistance with the hypervariable region analysis. Chromatography and mass spectrometry were performed in the Bioanalytical Facilities Core of the Massachusetts Institute of Technology Center for Environmental Health Sciences, supported by National Institute of Environmental Health Sciences Grant ES002109. This work was supported by National Science Foundation Grant CHE-1019990.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017261108/-/DCSupplemental.
References
- 1.Eckstein F, Gish G. Phosphorothioates in molecular biology. Trends Biochem Sci. 1989;14:97–100. doi: 10.1016/0968-0004(89)90130-8. [DOI] [PubMed] [Google Scholar]
- 2.Wang L, et al. Phosphorothioation of DNA in bacteria by dnd genes. Nat Chem Biol. 2007;3:709–710. doi: 10.1038/nchembio.2007.39. [DOI] [PubMed] [Google Scholar]
- 3.Zhou X, Deng Z, Firmin JL, Hopwood DA, Kieser T. Site-specific degradation of Streptomyces lividans DNA during electrophoresis in buffers contaminated with ferrous iron. Nucleic Acids Res. 1988;16:4341–4352. doi: 10.1093/nar/16.10.4341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ou HY, et al. dndDB: a database focused on phosphorothioation of the DNA backbone. PLoS ONE. 2009;4:e5132. doi: 10.1371/journal.pone.0005132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.You D, Wang L, Yao F, Zhou X, Deng Z. A novel DNA modification by sulfur: DndA is a NifS-like cysteine desulfurase capable of assembling DndC as an iron-sulfur cluster protein in Streptomyces lividans. Biochemistry. 2007;46:6126–6133. doi: 10.1021/bi602615k. [DOI] [PubMed] [Google Scholar]
- 6.Zhou X, et al. A novel DNA modification by sulphur. Mol Microbiol. 2005;57:1428–1438. doi: 10.1111/j.1365-2958.2005.04764.x. [DOI] [PubMed] [Google Scholar]
- 7.Yao F, Xu T, Zhou X, Deng Z, You D. Functional analysis of spfD gene involved in DNA phosphorothioation in Pseudomonas fluorescens Pf0-1. FEBS Lett. 2009;583:729–733. doi: 10.1016/j.febslet.2009.01.029. [DOI] [PubMed] [Google Scholar]
- 8.Hunt DE, et al. Resource partitioning and sympatric differentiation among closely related bacterioplankton. Science. 2008;320:1081–1085. doi: 10.1126/science.1157890. [DOI] [PubMed] [Google Scholar]
- 9.Wilson GG, Murray NE. Restriction and modification systems. Annu Rev Genet. 1991;25:585–627. doi: 10.1146/annurev.ge.25.120191.003101. [DOI] [PubMed] [Google Scholar]
- 10.Dyson P, Evans M. Novel post-replicative DNA modification in Streptomyces: analysis of the preferred modification site of plasmid pIJ101. Nucleic Acids Res. 1998;26:1248–1253. doi: 10.1093/nar/26.5.1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liang J, et al. DNA modification by sulfur: Analysis of the sequence recognition specificity surrounding the modification sites. Nucleic Acids Res. 2007;35:2944–2954. doi: 10.1093/nar/gkm176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ratel D, Ravanat JL, Berger F, Wion D. N6-methyladenine: The other methylated base of DNA. Bioessays. 2006;28:309–315. doi: 10.1002/bies.20342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kobayashi I. Behavior of restriction-modification systems as selfish mobile elements and their impact on genome evolution. Nucleic Acids Res. 2001;29:3742–3756. doi: 10.1093/nar/29.18.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.He X, et al. Analysis of a genomic island housing genes for DNA S-modification system in Streptomyces lividans 66 and its counterparts in other distantly related bacteria. Mol Microbiol. 2007;65:1034–1048. doi: 10.1111/j.1365-2958.2007.05846.x. [DOI] [PubMed] [Google Scholar]
- 15.Rusch DB, et al. The Sorcerer II Global Ocean Sampling expedition: Northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Morris RM, et al. SAR11 clade dominates ocean surface bacterioplankton communities. Nature. 2002;420:806–810. doi: 10.1038/nature01240. [DOI] [PubMed] [Google Scholar]
- 17.Treusch AH, et al. Seasonality and vertical structure of microbial communities in an ocean gyre. ISME J. 2009;3:1148–1163. doi: 10.1038/ismej.2009.60. [DOI] [PubMed] [Google Scholar]
- 18.Wolfe-Simon F, et al. A bacterium that can grow by using arsenic instead of phosphorus. Science. 2010 doi: 10.1126/science.1197258. in press. [DOI] [PubMed] [Google Scholar]
- 19.Xu T, Yao F, Zhou X, Deng Z, You D. A novel host-specific restriction system associated with DNA backbone S-modification in Salmonella. Nucleic Acids Res. 2010;38:7133–7141. doi: 10.1093/nar/gkq610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Olsen DB, Kotzorek G, Eckstein F. Investigation of the inhibitory role of phosphorothioate internucleotidic linkages on the catalytic activity of the restriction endonuclease EcoRV. Biochemistry. 1990;29:9546–9551. doi: 10.1021/bi00493a008. [DOI] [PubMed] [Google Scholar]
- 21.Olsen DB, Kotzorek G, Sayers JR, Eckstein F. Inhibition of the restriction endonuclease BanII using modified DNA substrates. Determination of phosphate residues critical for the formation of an active enzyme-DNA complex. J Biol Chem. 1990;265:14389–14394. [PubMed] [Google Scholar]
- 22.DuPont HL, et al. Pathogenesis of Escherichia coli diarrhea. N Engl J Med. 1971;285:1–9. doi: 10.1056/NEJM197107012850101. [DOI] [PubMed] [Google Scholar]
- 23.Levine MM, et al. Immunity to enterotoxigenic Escherichia coli. Infect Immun. 1979;23:729–736. doi: 10.1128/iai.23.3.729-736.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Murase T, Nagato M, Shirota K, Katoh H, Otsuki K. Pulsed-field gel electrophoresis-based subtyping of DNA degradation-sensitive Salmonella enterica subsp. enterica serovar Livingstone and serovar Cerro isolates obtained from a chicken layer farm. Vet Microbiol. 2004;99:139–143. doi: 10.1016/j.vetmic.2003.11.014. [DOI] [PubMed] [Google Scholar]
- 25.Lee HK, et al. Hahella chejuensis gen. nov., sp. nov., an extracellular-polysaccharide-producing marine bacterium. Int J Syst Evol Microbiol. 2001;51:661–666. doi: 10.1099/00207713-51-2-661. [DOI] [PubMed] [Google Scholar]
- 26.Pinhassi J, et al. Bermanella marisrubri gen. nov., sp. nov., a genome-sequenced gammaproteobacterium from the Red Sea. Int J Syst Evol Microbiol. 2009;59:373–377. doi: 10.1099/ijs.0.002113-0. [DOI] [PubMed] [Google Scholar]
- 27.Leonardo MR, et al. Shewanella pealeana sp. nov., a member of the microbial community associated with the accessory nidamental gland of the squid Loligo pealei. Int J Syst Bacteriol. 1999;49:1341–1351. doi: 10.1099/00207713-49-4-1341. [DOI] [PubMed] [Google Scholar]
- 28.Anderson RT, et al. Stimulating the in situ activity of Geobacter species to remove uranium from the groundwater of a uranium-contaminated aquifer. Appl Environ Microbiol. 2003;69:5884–5891. doi: 10.1128/AEM.69.10.5884-5891.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hopwood DA. Streptomyces in Nature and Medicine: The Antibiotic Makers. New York: Oxford Univ Press; 2007. [Google Scholar]
- 30.Rappé MS, Connon SA, Vergin KL, Giovannoni SJ. Cultivation of the ubiquitous SAR11 marine bacterioplankton clade. Nature. 2002;418:630–633. doi: 10.1038/nature00917. [DOI] [PubMed] [Google Scholar]
- 31.Giovannoni SJ, DeLong EF, Schmidt TM, Pace NR. Tangential flow filtration and preliminary phylogenetic analysis of marine picoplankton. Appl Environ Microbiol. 1990;56:2572–2575. doi: 10.1128/aem.56.8.2572-2575.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vergin KL, et al. High intraspecific recombination rate in a native population of Candidatus pelagibacter ubique (SAR11) Environ Microbiol. 2007;9:2430–2440. doi: 10.1111/j.1462-2920.2007.01361.x. [DOI] [PubMed] [Google Scholar]
- 33.Chaisson MJ, Brinza D, Pevzner PA. De novo fragment assembly with short mate-paired reads: Does the read length matter? Genome Res. 2009;19:336–346. doi: 10.1101/gr.079053.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zerbino DR, McEwen GK, Margulies EH, Birney E. Pebble and rock band: heuristic resolution of repeats and scaffolding in the velvet short-read de novo assembler. PLoS ONE. 2009;4:e8407. doi: 10.1371/journal.pone.0008407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Edgar RC. MUSCLE: Multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Guindon S, Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- 37.Letunic I, Bork P. Interactive Tree Of Life (iTOL): An online tool for phylogenetic tree display and annotation. Bioinformatics. 2007;23:127–128. doi: 10.1093/bioinformatics/btl529. [DOI] [PubMed] [Google Scholar]