Abstract
We tested the hypothesis that queen mandibular pheromone (QMP) causes changes in gene expression in the brain of the adult worker honey bee, and that these changes can be correlated to the downstream behavioral responses induced by QMP. In support of the first hypothesis, cage experiments revealed that QMP transiently regulated expression of several hundred genes and chronically regulated the expression of 19 genes. Several of these genes were also affected by QMP in experiments with bee colonies in the field, demonstrating robust gene regulation by pheromone. To evaluate the second hypothesis, we focused on one function of QMP: delaying the transition from working in the hive (e.g., brood care, or “nursing”) to foraging. We compared the list of QMP-regulated genes with the lists of genes differentially regulated in nurse and forager brains generated in a separate study. QMP consistently activated “nursing genes” and repressed “foraging genes,” suggesting that QMP may delay behavioral maturation by regulating genes in the brain that produce these behavioral states. We also report here on an ortholog of the Drosophila transcription factor kruppel homolog 1 that was strongly regulated by QMP, especially in the mushroom bodies of the bee brain. These results demonstrate chronic gene regulation by a primer pheromone and illustrate the potential of genomics to trace the actions of a pheromone from perception to action, and thereby provide insights into how pheromones regulate social life.
Many animal species, from insects to mammals, communicate via pheromones, chemicals that cause dramatic alterations in physiology and behavior. The recent identification of olfactory and pheromone receptors in Drosophila melanogaster and the mouse have provided new insights into how the olfactory system senses and encodes odorants (reviewed in refs. 1 and 2). It also has been demonstrated that pheromones affect gene expression in brain neurons, particularly with respect to immediate-early genes (3, 4). However, the molecular mechanisms by which pheromones are further transduced in the brain to influence behavior are only beginning to be understood (1). Particularly interesting are long-term changes in brain gene expression that might result from exposure to primer pheromones. These long-term changes in gene expression may be responsible for inducing long-term changes in physiology and behavior, a hallmark of primer pheromone action. Here we report on our efforts to use the honey bee (Apis mellifera) to study the effects of a primer pheromone on brain gene expression. We also have begun to correlate these gene expression changes with pheromone-mediated behavioral changes.
Honey bees show complex social organization that is controlled to a large extent by pheromones, many of which have been well characterized, both chemically and with respect to their specific behavioral effects (5–8). We studied the best understood bee pheromone, queen mandibular pheromone (QMP), a well-characterized blend that is part of a recently identified nine-component pheromone that attracts workers to attend the queen (9). QMP consists of five chemicals: (E)-9-keto-2-decenoic acid (9-ODA), (R,E)-(-)- and (S,E)-(+)-9-hydroxy-2-decenoic acid (9HDA), methyl p-hydroxybenzoate (HOB), and 4-hydroxy-3-methyoxyphenylethanol (HVA) (7). QMP plays many roles in social regulation (7). It prevents reproduction both by inhibiting worker ovary development (10) and the rearing of new queens (11). It may control brain development in the olfactory system of young bees (12). QMP also influences age-related division of labor among worker bees. Bees perform in-hive tasks such as brood care (nursing) for the first 2–3 weeks of their adult life and switch to foraging for food outside the hive in the last weeks of their life. Honey bees show many changes in physiology in association with this behavioral maturation, including structural changes in the mushroom bodies of the brain (which are sites of multimodal sensory integration and higher-order functions such as learning and memory), and differences in brain gene expression in nurses compared with foragers (13, 14). QMP delays honey bee behavioral maturation (15).
In this article we report on experiments that test two hypotheses: exposure to QMP causes changes in gene expression in the brain, and these changes correlate with the downstream behavioral effects of the pheromone. We used a recently developed honey bee brain microarray (16) to profile pheromone-induced changes in brain gene expression. Experiments were performed both with cage studies under controlled conditions in the laboratory and natural colony conditions in the field, to determine the robustness of the QMP effects. Because differences in gene expression associated with nursing and foraging behaviors have been analyzed extensively with microarrays (14), we used this information to explore our second hypothesis, that pheromone-regulated changes in gene expression are correlated with pheromone-induced changes in behavior. Finally, we began to characterize the effects of QMP on an ortholog of the Drosophila transcription factor kruppel homolog 1 (Kr-h1) (17), a particularly promising candidate gene that emerged from our microarray analyses.
Materials and Methods
Genetic Manipulation. Worker bees were derived from queens that were instrumentally inseminated with semen from a single, different drone, according to established procedures (18). Because male bees are haploid, the coefficient of relatedness among offspring of such an instrumentally inseminated queen is 0.75. Bees were obtained from three “source colonies,” each headed by one of these queens, R8, R11, or R16. Bees from “genotype” R8 were used for the microarray experiments, and bees from R11 and R16 were used to confirm some of the microarray results with real-time quantitative RT-PCR (qRT-PCR), as described below.
Rearing. Colonies were maintained at the University of Illinois Bee Research Facility according to standard commercial procedures. To provide bees of known age, honeycombs containing late-stage pupae were removed from source colonies and placed in an incubator to emerge (33°C, 95% relative humidity).
For cage studies, bees were collected 16 h after eclosion and placed in small (10 × 10 × 7 cm) Plexiglas cages (35 bees per cage) for 8 h before pheromone exposure began. Bees were provided with water and food (45% honey, 45% pollen, 10% water). Cages were kept in a humidity-controlled (50% relative humidity), dark incubator at 33°C. Pheromone exposure consisted of 0.1 queen equivalent (the typical amount found in one queen) of QMP (QMP+) or a solvent control (QMP-) introduced on a glass slide. This dose is sufficient for inhibiting ovary development in caged worker bees (10). The duration of pheromone exposure was 1, 2, 3, or 4 days. For each replicate, one entire QMP+ and one QMP- cage were flash-frozen in liquid nitrogen to prevent any additional changes in gene expression (19). Heads were removed and stored at -80°C until dissection. Bees were sampled at the same time of day to minimize any variation caused by circadian rhythms.
For field studies, the source colony was split into three colonies, allocating roughly equal quantities of adult bees, brood, and pollen and nectar stores. One colony retained the original queen (QR), one was left queenless (QL), and the third was given a strip that contained 10 queen equivalents (QMP+), a dose shown to mimic a live queen (20). The three colonies were transferred to a different apiary >2 miles away so they would not return to the site of their natal hive. Before the colony split, bees (n ≈1,500) were collected 0–36 h after eclosion, marked on the dorsal thorax with a paint dot (Testor's Paint, Rockford, IL), and ≈500 were placed in each of the three colonies. Two days later, the marked bees were collected (n = 100). Bees were collected into liquid nitrogen, and heads were stored as above.
For gene expression analysis in the mushroom bodies, foragers were collected from two colonies (R5, R11) upon return from foraging flights, and 1-day-old bees were collected <24 h after eclosion.
Pheromones. QMP was obtained from PheroTech (Delta, Canada). For cage experiments, 0.1 QMP was applied to glass slides in 10 μl of 1% H2O/isopropyl alcohol and allowed to dry (9). QMP was applied to a fresh slide at the same time every day. For field experiments, commercially formulated QMP strips (Phero-Tech) were used and replaced daily (20).
Confirmation of Pheromonal Activity. To confirm QMP activity in cages, the effect of QMP on ovary development was assessed (10). Cages of bees were set up as described above and maintained for 10 days, then collected and stored at -20°C until dissection. The degree of ovary development was assessed according to Veltuis (21). Two cages from each of the three genotypes were analyzed. A total of 12 of 30 QMP- bees developed ovaries (with developed eggs and follicle cells), whereas 0 of the 30 QMP+ bees developed ovaries (P < 0.01, χ2 test), demonstrating that the QMP was active and the bees were sensitive to it.
To confirm QMP activity in the field, the effect of QMP on the number of queen cells built was determined. Queen cells are distinctive large cells on the honeycomb built by worker bees to rear new queens (8); QMP partially inhibits this behavior (11). The number of queen cells built in QL colonies was 31, 29, and 12 (R8, R16, and R11, respectively) compared with 1, 5, and 5 queen cells built in the QMP+ colonies. In QR colonies, no queen cells were built in R8 and R16, whereas three were built in R11. QMP clearly suppressed queen cell-building behavior in the field.
Brain Dissection. Whole bee heads were partially lyophilized to facilitate brain dissection (22). Dissections were performed over dry ice so tissue never thawed. Because ocelli and the subesophageal ganglion frequently fractured during dissection, these were removed during all dissections, while the remainder of the brain was included. Mushroom bodies were dissected from freeze-dried brains as in ref. 22.
Microarrays and Data Analysis. The microarray contained ≈9,000 cDNAs, representing ≈7,600 different genes, 40% of which have been annotated primarily by using comparisons to Drosophila genes and the molecular function classification scheme of the Gene Ontology Consortium (16). This number is estimated to account for 50% of the genes in the honey bee genome, based on comparisons with the Drosophila genome.
For cage studies, direct competitive hybridization comparisons were made for matched samples of QMP+ and QMP- bees. Each sample consisted of 10 brains pooled (10 bees taken randomly from a cage of 35 bees after the entire cage was killed). Eight arrays were analyzed for each time point (1, 2, 3, or 4 days of pheromone exposure), a total of 32 microarrays. The eight replicates were comprised of four biological replicates (different cages) and four technical replicates. For field studies, each sample consisted of 10 brains pooled. Samples from the QR, QMP+, and QL colonies split from the R8 source colony were compared with each other in a loop design that used three microarrays, with four technical replicates, for a total of 12 arrays (23).
Dissected bee brains were homogenized in Trizol (Invitrogen Life Technologies), and RNA was extracted according to the manufacturer's protocols. mRNA was then purified by using an Oligotex mRNA kit (Qiagen, Valencia, CA). cDNA synthesis and hybridization to microarrays followed protocols modified from ref. 24 and described in ref. 14. Arrays were scanned with GENEPIX software and normalized to the median intensity with regional and intensity-dependent Lowess normalization. Dyes used to label the QMP+ and QMP- samples were reversed in half of the replicates to control for dye-by-gene interactions. The following filtering protocols were followed, as in Whitfield et al. (14). cDNAs with average expression intensities across each set of eight arrays <350 or absent from ≥1 array were removed from the analysis. cDNAs expressed at equal levels in hypopharyngeal glands in the honey bee head (14) were removed because material from these glands can sometimes contaminate dissected brains. Only cDNAs expressed at all time points and also in the Whitfield et al. (14) nurse vs. forager analysis were analyzed, to facilitate comparisons across the two studies. This left 6,360 cDNAs, representing ≈5,000 unique genes.
Bayesian analysis (25) was used to determine which cDNAs were significantly regulated by QMP. This method generates an estimated mean and 95% confidence interval (CI) for the relative level of expression of each cDNA for each experimental group of bees. cDNAs were classified as differentially expressed if their 95% CIs did not overlap between groups (one-tailed t test). This approach enables identification of small, but reproducible, differences in gene expression (26, 27). Notably, the expression of most significantly regulated genes changed by ≈10%. For example, of 2,607 cDNAs found to be significantly regulated during the time course, only 1,158 showed differences of >10%, and only 111 showed differences of >20%. mRNA Quantification by Real-Time qRT-PCR. Confirmation of some of the results obtained from microarray analysis was performed with real-time qRT-PCR in individual brains (19) with an ABI Prism 7900 sequence detector and the SYBR green detection method (Applied Biosystems). Rp49 or eIF3-S8, two housekeeping genes that did not vary in expression levels on these microarrays, were used as loading controls. Quantification was as described (19). The sequences for the primers used are given in Table 4, which is published as supporting information on the PNAS web site.
Results
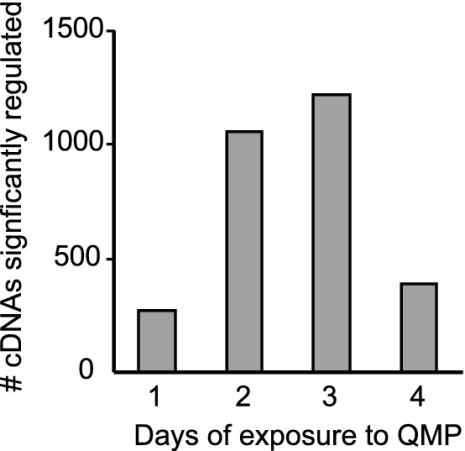
QMP Influences Brain Gene Expression in the Laboratory. There were 1,210 cDNAs with significantly higher expression and 1,397 cDNAs with significantly lower expression in the brains of QMP+ bees relative to QMP- bees. This is about eight times more than the number of false positives expected (25). Several hundred cDNAs were significantly regulated on each day of the 4-day time course, with maximum numbers on days 2 and 3 and the fewest on day 1 (Fig. 1). The magnitude differences were smallest on day 1 as well: expression of only 3 ESTs changed by 40% on day 1, whereas 21, 20, and 21 ESTs, respectively, were regulated by 40% on the other three days. Expression levels for most cDNAs changed during the time course, but a small subset was chronically regulated across days 2, 3, and 4 (13 up-regulated and 6 down-regulated). Eight of these chronically regulated genes are annotated, with high sequence similarity to known Drosophila or human genes (Table 1). They include the transcription factor Kr-h1 (28), the G protein-coupled receptor frizzled 2 (29), and a transmembrane protein involved in regulating glial cell thickness (push/poe) (30). See Table 5, which is published as supporting information on the PNAS web site, for a complete list of regulated genes.
Fig. 1.
Dynamic regulation of gene expression in the honey bee brain by QMP. Bees were maintained in cages in the laboratory either with or without QMP. The number of cDNAs showing significant differences in expression on each day is shown.
Table 1. Annotated genes chronically regulated by QMP in the honey bee brain.
| Closest Drosophila (or human) match | Similarity score | Fold difference QMP+/QMP- | Possible function | Nurse/forager |
|---|---|---|---|---|
| Q9H2Y7 (human) | 2E-08 | 1.11 | WD-repeat protein | Nurse |
| CG14168 | 1E-13 | 1.12 | PDZ domain | Nurse |
| clt | 5E-23 | 1.16 | Carboxylesterase | |
| Traf1 | 1E-24 | 1.16 | Tumor necrosis factor receptor-associated factor | Forager |
| CG7474 | 4E-85 | 1.17 | Tubulinyl-tyrosine ligase | |
| poe | 9E-34 | 0.86 | Calmodulin binding, synpatogenesis | |
| frizzled 2 | 5E-96 | 0.70 | G protein-coupled receptor | Forager |
| kr-h1 | 1E-100 | 0.59 | Zn-finger transcription factor | Forager |
Genes found to be chronically regulated by QMP with high sequence similarity to known Drosophila or human genes are listed below. Similarity score: BLAST E value (this corresponds to the EST sequency only, except for Kr-hl, for which full sequence was obtained, and frizzled 2, for which additional sequence was identified from the genome project, http://hgsc.bcm.tmc.edu/blast/?organism=Amellifera). Fold difference column: the maximum difference between values obtained for bees maintained in cages with or without QMP. Nurse/forager column indicates in which behavioral group the gene was found to be significantly upregulated in an independent microarray-based study (14).
QMP-Mediated Gene Expression Correlates with Downstream Behavioral Effects. If QMP regulation of gene expression in the brain is related to the QMP-mediated delay in the transition from hive work to foraging (15), then the following pattern is predicted: exposure to QMP activates genes in the brain associated with nursing and represses genes associated with foraging. The prediction was tested by comparing our data with results from a previous study (14). That study showed widespread differences in gene expression between nurses and foragers, with 1,360 showing significantly more mRNA abundance in nurse brains (“nurse list”) and 1,310 significantly more mRNA abundance in forager brains (“forager list”). Results of this comparison agree with the prediction. A significantly larger proportion of QMP-up-regulated cDNAs was on the nurse list than were QMP-down-regulated cDNAs (Table 2). Likewise, a significantly larger proportion of QMP-down-regulated cDNAs were on the forager list than were QMP-up-regulated cDNAs. (Table 2). These trends were even more striking if only the chronically regulated cDNAs are considered (Table 2). See Table 5 for a complete listing of all QMP-regulated genes that are also on the nurse or foragers lists.
Table 2. QMP activates genes in the bee brain associated with nursing and represses genes associated with foraging.
| Location | QMP | No. of cDNAs | % on nurse list | % on forager list |
|---|---|---|---|---|
| Cage | Total QMP ↓ | 1,397 | 14 | 27 |
| Total QMP ↑ | 1,210 | 33 | 12 | |
| Chronically ↓ | 6 | 0 | 67 | |
| Chronically ↑ | 13 | 62 | 8 | |
| Colony | QMP ↓ | 374 | 13 | 26 |
| QMP ↑ | 323 | 42 | 16 | |
| Cage and colony | QMP ↓ | 59 | 15 | 42 |
| QMP ↑ | 74 | 53 | 3 |
QMP-regulated cDNAs were compared to the lists of cDNAs significantly up-regulated in nurse bees or up-regulated in foragers. A significantly larger proportion of QMP ↑ (up-regulated) cDNAs were on the nurse list than were QMP ↓ cDNAs. Likewise, a significantly larger proportion of QMP ↓ cDNAs were on the forager list than were QMP ↑ cDNAs (P < 0.001, χ2 tests). Genes chronically regulated by QMP showed the same trends.
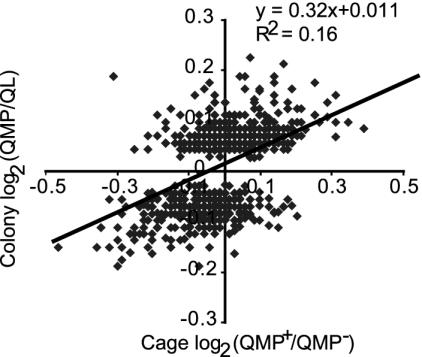
QMP Influences Brain Gene Expression in Bee Colonies. There were 697 cDNAs that showed significant differences in brain expression in bees sampled from QMP+ and QL colonies (Fig. 2). Overall, these expression differences were most similar to the results from cages sampled on day 3. For example, of the cDNAs significantly up-regulated in the field, 59 were also significantly up-regulated in the cages (vs. 20 that were down-regulated in the cages), and of the cDNAs down-regulated in the field, 74 were down-regulated in cages (vs. 7 that were up-regulated in the cages). Relative to cage bees, there were fewer cDNAs with significant differences in colony bees, and they showed expression differences of smaller magnitude. Among the genes significantly regulated in the brain by QMP in both cage and colony were: transcription factors HLH3B and klumpfuss, gap junction proteins inx2 and inx3, Tor, a phosphatidylinositol 3-kinase, Nrv2, an Na+/K+ exchanging ATPase complex, RacGAP, a GTPase activator, and Rab6, a GTPase.
Fig. 2.
Comparison of QMP-regulated brain gene expression in bees from colonies in the field vs. cages in the laboratory. The cDNAs showing significant differences in expression in bees from QMP vs. QL (queenless) colonies are shown (minus three outliers, n = 694). The expression levels (log-transformed) of these cDNAs under colony and cage (day 3) conditions are plotted, and the regression line is shown. In general, cDNAs down-regulated in colony bees were down-regulated in cage bees, and likewise for up-regulated cDNAs. Expression differences were generally lower in colony bees.
As was the case in the laboratory experiments, a significantly larger proportion of QMP-up-regulated cDNAs were on the nurse list than were QMP-down-regulated cDNAs, and likewise for QMP-down-regulated cDNAs on the forager list (Table 2). These trends were even more striking if only the cDNAs regulated in both the cages and colonies are considered (Table 2).
The field experiments also allowed us to compare the effects of a live queen to the effects of QMP on gene expression in the brain. A total of 1,047 cDNAs were differentially expressed between the QR and QMP colonies; thus, QMP did not completely mimic the live queen. A total of 335 cDNAs were coregulated in the QR and QMP colonies relative to the QL colony; these genes may be specifically regulated by QMP released by the queen.
QMP Effects on Transcription Factor. The gene with the biggest difference in mean expression levels in the cage experiments was a transcription factor, Kr-h1. Because transcription factors regulate the expression of other genes, they may serve as markers for regulation of pheromone-mediated “transcriptional programs” that control downstream behavioral effects. A total of 39 of the 129 cDNAs annotated as transcription factors (16) were significantly regulated by exposure to QMP (Table 6, which is published as supporting information on the PNAS web site), whereas 17 were significantly regulated by >10%. This proportion was relatively high compared with other functional categories of genes (Table 3).
Table 3. Effects of QMP on different functional categories of genes.
| Functional category | No. annotated | % Regulated |
|---|---|---|
| Protein phosphatases | 35 | 2.9 |
| Synaptic vesicle transport | 66 | 1.5 |
| Ion channels | 73 | 6.8 |
| Protein kinases | 87 | 8.0 |
| Cytoskeletal organization and biogenesis | 89 | 4.5 |
| Receptors | 106 | 5.7 |
| Peptidases | 108 | 5.6 |
| RNA-binding proteins | 127 | 3.9 |
| Oxidoreductases | 129 | 8.5 |
| Transcription factors | 129 | 13.2 |
For each functional category, the number of genes significantly regulated by QMP (>10%) for at least 1 day in cage experiments are shown. Categories of molecular function are from the Gene Ontology Consortium; annotation is from ref. 16.
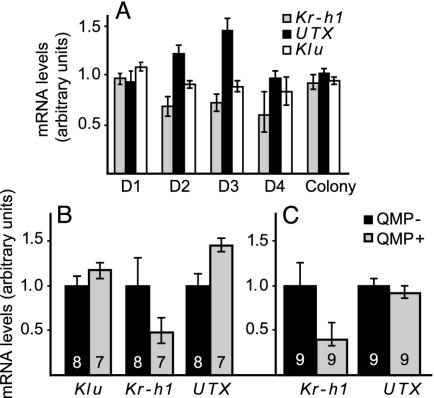
Microarray analysis showed that brain expression of three transcription factors (Kr-h1, Utx, and klumpfuss) was significantly affected by QMP over more than 1 day (Fig. 3A). In the colony experiments, only Klumpfuss was significantly regulated. The cage results were verified with qRT-PCR, using individual bee brains rather than samples of pooled brains (Fig. 3 B and C). In genotype R8 (Fig. 3B), which was the same as that used for the microarray studies, brain expression of Kr-h1 and Utx was significantly decreased and increased, respectively, in QMP+ bees, as in the microarray analyses, whereas klumpfuss levels were not significantly different. In a second genotype (Fig. 3C), Kr-h1 brain expression again was significantly lower in QMP+ bees, but there was no effect of QMP on Utx expression levels. Because Utx was up-regulated for only 2 days in the microarray experiments, it is possible that its temporal pattern of QMP-mediated expression was different in this genotype. Regardless, Kr-h1 clearly showed the most robust and consistent pattern of QMP regulation.
Fig. 3.
Confirmation of microarray results. mRNA levels for three transcription factors (kr-h1, utx, and klu) shown by microarray analysis to be significantly regulated by QMP were quantified by real-time qRT-PCR in individual brains. (A) Expression levels (QMP+/QMP- ratio) determined by microarray analysis on days (D) 1–4 for cage bees and colony bees. (B) Expression levels determined by qRT-PCR for bees exposed to QMP for 3 days from the same genotype as in A. Number of individually analyzed brains is indicated on bars. Data are means ± SE (converted to the same arbitrary scale as the mean), normalized to the QMP- sample. Statistical analysis was done by one-tailed t test. (C) Same as B, except bees were from a second genotype.
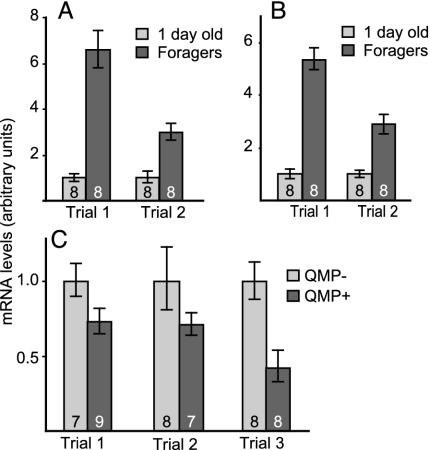
Two additional experiments were conducted to further establish Kr-h1 as a candidate gene to study long-term, behaviorally relevant changes in brain gene expression caused by pheromone exposure. First, we determined whether QMP effects on Kr-h1 expression in the brain were associated with behaviorally related differences in expression. Previous microarray studies indicated that Kr-h1 expression is higher in foragers relative to nurses (14). qRT-PCR analysis agreed, showing that brain Kr-h1 expression was increased in foragers relative to 1-day-old bees (Fig. 4A). Second, we determined whether Kr-h1 is expressed in the mushroom bodies, as a first step toward exploring the effects of pheromone on gene expression in brain regions involved in higher-order integration and processing. Kr-h1 was expressed in the mushroom bodies, with expression significantly higher in foragers relative to 1-day-old bees (Fig. 4B) and also significantly higher in QMP- bees than QMP+ bees (Fig. 4C). Kr-h1 expression levels were not significantly different in optic lobes of QMP- vs. QMP+ bees (data not shown), suggesting that expression of this gene is specifically regulated in the mushroom bodies rather than the whole brain. Cloning A. mellifera kr-h1 (GenBank accession no AY338499, see Fig. 5, which is published as supporting information on the PNAS web site) revealed that it contains eight zinc finger domains from amino acids 62–289 and has significant sequence similarity to the Drosophila Kr-h1 protein and the partial Anopheles gambiae protein. There are two transcripts for Kr-h1 present in Drosophila, Kr-h1 RA and RB (28). The RA transcript is present in larvae and adults, whereas the RB transcript is present only in embryos and has an additional 55 aa on the N terminal. The cloned A. mellifera kr-h1 does not contain this N-terminal extension, and thus matches the RA transcript most closely. However, it is ≈200 aa shorter than the RA, because of deletions throughout the gene.
Fig. 4.
Behavior- and QMP-related Kr-h1 expression in whole brains and mushroom bodies in the honey bee. mRNA levels of Kr-h1 in 1-day-old bees and foragers were quantified with qRT-PCR in whole brains (A) and mushroom bodies (B) in two genotypes (trials 1 and 2). Differences were significant (P < 0.001, one-tailed t test). (C) Effect of QMP on Kr-h1 mRNA levels in mushroom bodies. Differences were significant in trial 1 (P < 0.05) and trial 3 (P < 0.01) but not in trial 2 (P = 0.09). Other notations are as in Fig. 3.
Discussion
This study has demonstrated that QMP causes changes in expression levels of many genes in the brain of adult honey bees, and that these changes correlate with some of the downstream behavioral effects of the pheromone. QMP effects on gene expression were detected both in a controlled laboratory environment and in bee colonies in the field, which represent a more natural environment. QMP causes transient changes in expression of several hundred genes, but causes chronic changes in only a small subset of genes in the brain.
The effects of QMP on brain gene expression changed over time. One possible explanation for this is that the transcriptional response to QMP is dynamic, with one set of genes activating/ repressing expression of another downstream set of genes in “transcriptional cascades” reminiscent of what occurs during Drosophila neural development (31). These genes may only need to act transiently to produce the necessary response to queen pheromone, such as neuronal remodeling of pheromone-responsive networks. For example, bees raised in queenless conditions for 4 days have smaller synaptic structures in the antennal lobe, suggesting that QMP may influence synaptic growth and remodeling during this time frame (12). Thus, the dynamic nature of the response to QMP might be related to the fact that the bee's olfactory system continues developing for the first few days of adult life (32).
Alternatively, it simply might be difficult to detect small, but statistically significant, differences in gene expression across consecutive days, even with the relatively large number of replicates used in this study. In most cases, the observed significant changes in gene expression were relatively subtle (<20%). Perhaps the relatively small QMP-induced expression differences are caused by effects of QMP on a small set of neurons. Small, but statistically significant, differences in gene expression are detectable in microarrays studies such as ours with numerous replicates (14, 27) and can have biological significance (26, 27).
Some genes chronically regulated by QMP may be involved in stably altering neuronal activity and responsiveness. For example, one of the chronically down-regulated genes encodes a homolog of the Drosophila protein Pushover (also named Poe), which has been shown to play a role in regulating glial cell thickness (30) and neuronal excitability (33). Such alterations in central brain processing regions could lead to changes in responsiveness to various stimuli, thereby altering behavior (34).
There was considerable overlap in the genes regulated by QMP in the cages and in colonies, but gene expression changes in the colonies were smaller and involved fewer genes. This might be because the “QMP strip” used in the colony releases a lower amount of pheromone per bee than the glass slide in the cages; thus, the colony bees may have been exposed to an almost 2-fold lower dose of QMP (K. N. Slessor, personal communication). Another factor is that the colony environment is more varied with respect to pheromonal and other stimuli. In particular, the brood in the colonies release a pheromone that produces many of the same effects as QMP (35–38), and this may have attenuated the response to QMP in the colony experiment. If so, then the set of genes that were found to be regulated in both the colony and the cage experiments may be robustly regulated by or only sensitive to QMP. Further cage experiments using brood pheromone can help address these questions.
QMP and a live queen had similar effects on some, but not all, genes. This finding is consistent with observations showing that QMP is not as effective as a full queen extract in producing behavioral and physiological responses. For example, an additional four components were recently identified that improve the performance of synthetic queen pheromone in the attraction of workers to the queen, and even this nine-component blend is not quite as effective as the full extract (9). The genes that were specifically regulated only under queenright conditions may be responding to other unidentified components of queen pheromone and may prove to be reliable markers for future elucidation of a more complete pheromone blend.
QMP consistently activated genes correlated with nursing behavior and repressed genes correlated with foraging behavior, in both cage and colony experiments. This finding suggests that the effects of QMP on the timing of the transition from working in the hive to foraging (15) may be caused by QMP regulation of genes in the brain that produce these behavioral states. At least some QMP-regulated genes may be involved in regulating the transitions to nursing and foraging, rather than the maintenance of these states, because the bees we used were too young or simply unable to perform these behaviors in laboratory cages. The fact that many QMP-regulated genes were not found to be associated with nursing or foraging behavior may reflect the fact that QMP also is involved in the regulation of several other behavioral and physiological processes besides age-related division of labor, such as inhibiting ovary development and the rearing of new queens. Further microarray experiments may identify distinct transcriptional programs that relate to other pheromone-regulated processes.
The proportion of transcription factors regulated by QMP was relatively high compared with other functional groups of genes. This result suggests transcription factors may be important targets of pheromone activation. To aid in the future identification of specific transcriptional programs, we catalogued the transcription factors whose expression levels were significantly regulated by QMP. Only one transcription factor was found to be chronically regulated; most showed significant changes in expression on only 1 day. Transiently regulated transcription factors may simply initiate a downstream program and be subsequently turned off in a matter of hours as is the case for CREB (cyclic AMP-response element binding protein), which initiates changes in neuronal plasticity (reviewed in refs. 39 and 40).
Kr-h1 was the most highly and robustly regulated gene identified in this study. Furthermore, Kr-hl has the unique trait of being chronically regulated by a pheromone. Kruppel homologs are zinc finger transcription factors that play important roles in orchestrating development and cell differentiation (reviewed in (17), including neural development (31). Kr-h1 in particular was identified as an ecdysone-sensitive transcript during Drosophila morphogenesis (28), and in a gain-of-function screen for genes involved in motor axon guidance and synaptogenesis in Drosophila larvae (41). Differences in expression patterns and sequence divergences within the DNA-binding regions of Kruppel-like proteins across mammalian lineages suggest that members of this family have undergone duplication and acquired novel functions in different species during evolution (42).
The A. mellifera Kr-h1 protein is highly similar to Drosophila Kr-h1 and the Anopheles gambaie ortholog, suggesting that it functions similarly in all three insects. However, in the case of the honey bee, a species with a highly derived form of social organization, it has evolved to be regulated by a pheromone, perhaps in addition to intrinsic factors common to the dipteran species with solitary lifestyles. QMP regulation of Kr-h1 occurs in the mushroom bodies, the sites of integration of sensory information, so Kr-h1 may be involved in organizing stable changes in gene expression and neuron structure that are necessary to transduce the chemosensory QMP stimulus to downstream changes in behavior and physiology. Further experiments will be necessary to map Kr-h1 expression throughout the brain to further elucidate the function of Kr-h1 in response to pheromone. Given that Kr-h1 is strongly down-regulated by QMP, it is puzzling that Kr-h1 is repressed by QMP in young honey bees but up-regulated in older bees, because both groups were taken from colonies with a queen. Perhaps this is because foragers typically contact the queen less than do younger bees, or foragers are simply less responsive to QMP. Further experiments will be needed to explore this issue.
QMP affects the expression of many genes in the bee brain, and in particular changes expression of genes associated with behaviors that also are regulated by this pheromone. Some effects on gene expression are relatively transient, which may reflect short-term modifications in synaptic plasticity, and some are more chronic, which may reflect long-term changes in neuronal responsiveness and behavioral state. Some of the genes identified here may represent transcriptional programs regulating particular pheromone-mediated physiological and behavioral processes. Further genomewide analysis of the transcriptional effects of QMP and other pheromones should lead to new insights into how pheromones regulate social life.
Supplementary Material
Acknowledgments
We thank K. Pruiett for expert assistance with the bees; A. Barron, C. Schook, and L. Wraight for technical assistance; R. Velarde for samples of 1-day-old and forager bees; S. Rodriguez-Zas for help with statistical analysis; M. Band for fabricating microarrays of outstanding quality; C. Keeling, S. Hoover, K. N. Slessor, and M. L. Winston for helpful advice; and R. Maleszka and members of the Robinson laboratory for comments that improved the manuscript. This work was supported by a Beckman Institute postdoctoral fellowship (to C.M.G.), a National Science Foundation Bioinformatics postdoctoral fellowship (to C.W.W.), and grants from the Burroughs Welcome Trust, the National Institutes of Health, and the U.S. Department of Agriculture (to G.E.R.).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Chemical Communication in a Post-Genomic World,” held January 17–19, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: QMP, queen mandibular pheromone; Kr-h1, kruppel homolog 1; qRT-PCR, quantitative RT-PCR.
Data deposition: Gene expression data meet Minimum Information About a Microarray Experiment (MIAME) standards and have been deposited at ArrayExpress (www.ebi.ac.uk/arrayexpress).
References
- 1.Dulac, C. & Torello, A. T. (2003) Nat. Rev. Neurosci. 4, 551-562. [DOI] [PubMed] [Google Scholar]
- 2.Vosshall, L. B. (2001) Chem. Senses 26, 207-213. [DOI] [PubMed] [Google Scholar]
- 3.Halem, H. A., Baum, M. J. & Cherry, J. A. (2001) J. Neurosci. 21, 2474-2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brennan, P. A., Schellinck, H. M. & Keverne, E. B. (1999) Neuroscience 90, 1463-1470. [DOI] [PubMed] [Google Scholar]
- 5.Free, J. B. (1987) Pheromones of Social Bees (Cornell Univ. Press, Ithaca, NY).
- 6.Slessor, K. N., Kaminski, L.-A., King, G. G. S., Borden, J. H. & Winston, M. L. (1988) Nature 332, 354-356. [Google Scholar]
- 7.Winston, M. L. & Slessor, K. N. (1998) Apidologie 29, 81-95. [Google Scholar]
- 8.Winston, M. L. (1987) The Biology of the Honey Bee (Harvard Univ. Press, Cambridge, MA).
- 9.Keeling, C. I., Slessor, K. N., Higo, H. A. & Winston, M. L. (2003) Proc. Natl. Acad. Sci. USA 100, 4486-4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hoover, S. E. R., Keeling, C. I., Winston, M. L. & Slessor, K. N. (2003) Naturwissenschaften 90, 477-480. [DOI] [PubMed] [Google Scholar]
- 11.Winston, M. L., Higo, H. A., Colley, S. J., Pankiw, T. & Slessor, K. N. (1991) J. Insect Behav. 4, 649-660. [Google Scholar]
- 12.Morgan, S. M., Butz Huryn, V. M., Downes, S. R. & Mercer, A. R. (1998) Behav. Brain Res. 91, 115-126. [DOI] [PubMed] [Google Scholar]
- 13.Robinson, G. (2002) Am. Nat. 160, S160-S172. [DOI] [PubMed] [Google Scholar]
- 14.Whitfield, C. W., Cziko, A.-M. & Robinson, G. (2003) Science 302, 296-299. [DOI] [PubMed] [Google Scholar]
- 15.Pankiw, T., Huang, Z.-Y., Winston, M. L. & Robinson, G. E. (1998) J. Insect Physiol. 44, 685-692. [DOI] [PubMed] [Google Scholar]
- 16.Whitfield, C. W., Band, M. R., Bonaldo, M. F., Kumar, C. G., Liu, L., Pardinas, J. R., Robertson, H. M., Soares, M. B. & Robinson, G. E. (2002) Genome Res. 12, 555-566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kaczynski, J., Cook, T. & Urrutia, R. (2003) Genome Biol. 4, 206.1-206.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laidlaw, J., H. H. (1987) Bee World, 17-38.
- 19.Ben-Shahar, Y., Robichon, A., Sokolowski, M. B. & Robinson, G. E. (2002) Science 296, 741-744. [DOI] [PubMed] [Google Scholar]
- 20.Ledoux, M. N., Winston, M. L., Higo, H. A., Keeling, C. I., Slessor, K. N. & Le Conte, Y. (2001) Insectes Soc. 48, 14-20. [Google Scholar]
- 21.Veltuis, H. H. W. (1970) Entolmol. Exp. Appl. 13, 377-394. [Google Scholar]
- 22.Schulz, D. J. & Robinson, G. E. (1999) J. Comp. Physiol. A 184, 481-488. [DOI] [PubMed] [Google Scholar]
- 23.Kerr, M. K. & Churchill, G. A. (2001) Genet. Res. 77, 123-128. [DOI] [PubMed] [Google Scholar]
- 24.Hegde, P., Qi, R., Abernathy, K., Gay, C., Dharap, S., Gaspard, R., Hughes, J. E., Snesrud, E., Lee, N. & Quackenbush, J. (2000) BioTechniques 29, 548-550, 552-554, 556. [DOI] [PubMed] [Google Scholar]
- 25.Townsend, J. P. & Hartl, D. L. (2002) Genome Biol. 3, 0071.1-0071.6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yan, X. D., Hanson, A. J., Nahreini, P., Koustas, W. T., Andreatta, C. & Prasad, K. N. (2002) In Vitro Cell Dev. Biol. Anim. 38, 529-537. [DOI] [PubMed] [Google Scholar]
- 27.Gibson, G. (2002) Mol. Ecol. 11, 17-24. [DOI] [PubMed] [Google Scholar]
- 28.Pecasse, F., Beck, Y., Ruiz, C. & Richards, G. (2000) Dev. Biol. 221, 53-67. [DOI] [PubMed] [Google Scholar]
- 29.Bhanot, P., Brink, M., Samos, C. H., Hsieh, J. C., Wang, Y., Macke, J. P., Andrew, D., Nathans, J. & Nusse, R. (1996) Nature 382, 225-230. [DOI] [PubMed] [Google Scholar]
- 30.Yager, J., Richards, S., Hekmat-Scafe, D. S., Hurd, D. D., Sundaresan, V., Caprette, D. R., Saxton, W. M., Carlson, J. R. & Stern, M. (2001) Proc. Natl. Acad. Sci. USA 98, 10445-10450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Isshiki, T., Pearson, B., Holbrook, S. & Doe, C. Q. (2001) Cell 106, 511-521. [DOI] [PubMed] [Google Scholar]
- 32.Pham-Delengue, M. H., Trouiller, J., Caillaud, C. M., Roger, B. & Masson, C. (1993) Apidologie 24, 267-281. [Google Scholar]
- 33.Richards, S., Hillman, T. & Stern, M. (1996) Genetics 142, 1215-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Beshers, S. N., Robinson, G. E. & Mittenthal, J. E. (1999) in Information Processing in Social Insects, ed. Detrain, C. (Birkhäuser, Boston), pp. 115-139.
- 35.Arnold, G., Le Conte, Y., Trouiller, J., Hervet, H., Chappe, B. & Masson, C. (1994) C. R. Acad. Sci. Ser. III 317, 511-515. [Google Scholar]
- 36.Le Conte, Y., Areski, M. & Robinson, G. E. (2001) Proc. R. Soc. London Ser. B 268, 1-6. [Google Scholar]
- 37.Mohammedi, A., Crauser, D., Paris, A. & Le Conte, Y. (1996) C. R. Acad. Sci. Ser. III 319, 769-772. [PubMed] [Google Scholar]
- 38.Mohammedi, A., Paris, A., Crauser, D. & Le Conte, Y. (1998) Naturwissenschaften 85, 455-458. [Google Scholar]
- 39.Clayton, D. F. (2000) Neurobiol. Learn. Mem. 74, 185-216. [DOI] [PubMed] [Google Scholar]
- 40.West, A. E., Griffith, E. C. & Greenberg, M. E. (2002) Nat. Rev. Neurosci. 3, 921-931. [DOI] [PubMed] [Google Scholar]
- 41.Kraut, R., Menon, K. & Zinn, K. (2001) Curr. Biol. 11, 417-430. [DOI] [PubMed] [Google Scholar]
- 42.Shannon, M., Hamilton, A. T., Gordon, L., Branscomb, E. & Stubbs, L. (2003) Genome Res. 13, 1097-1110. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.