Abstract
Repeated cocaine exposure induces persistent alterations in genome-wide transcriptional regulatory networks, chromatin remodeling activity and, ultimately, gene expression profiles in the brain's reward circuitry. Virtually all previous investigations have centered on drug-mediated effects occurring throughout active euchromatic regions of the genome, with very little known concerning the impact of cocaine exposure on the regulation and maintenance of heterochromatin in adult brain. Here, we report that cocaine dramatically and dynamically alters heterochromatic histone H3 lysine 9 trimethylation (H3K9me3) in the nucleus accumbens (NAc), a key brain reward region. Furthermore, we demonstrate that repeated cocaine exposure causes persistent decreases in heterochromatization in this brain region, suggesting a potential role for heterochromatic regulation in the long-term actions of cocaine. To identify precise genomic loci affected by these alterations, chromatin immunoprecipitation followed by massively parallel DNA sequencing (ChIP-Seq) was performed on NAc. ChIP-Seq analyses confirmed the existence of the H3K9me3 mark mainly within intergenic regions of the genome and identified specific patterns of cocaine-induced H3K9me3 regulation at repetitive genomic sequences. Cocaine-mediated decreases in H3K9me3 enrichment at specific genomic repeats [e.g., long interspersed nuclear element (LINE)-1 repeats] were further confirmed by the increased expression of LINE-1 retrotransposon-associated repetitive elements in NAc. Such increases likely reflect global patterns of genomic destabilization in this brain region after repeated cocaine administration and open the door for future investigations into the epigenetic and genetic basis of drug addiction.
Repeated cocaine administration promotes long-lasting changes in gene expression within the nucleus accumbens (NAc), a key component of the brain's reward circuitry. Such gene expression changes are believed to contribute to persistent alterations in behavior associated with cocaine addiction (1). Increasing evidence supports an important role for chromatin regulation in promoting aberrant transcriptional changes in this brain region that may underlie aspects of addiction (2–9). Numerous posttranslational modifications of histones, such as acetylation, phosphorylation, and methylation, have been shown to modulate gene transcription in diverse tissues via regulation of chromatin packaging and recruitment of effector proteins (10). These interdependent modifications, which occur at multiple positions throughout the N termini of histone proteins, provide marks for repressed vs. active chromatin.
Recent work has demonstrated that cocaine regulates chromatin structure in NAc, in part, through direct cocaine-induced perturbations of histone-modifying enzymes, as evidenced by decreased expression and activity of the histone methyltransferase G9a, which leads to globally decreased repressive histone methylation, enhanced euchromatic gene expression, and sensitized behavioral and structural plasticity after repeated cocaine administration (9). Alterations in histone acetyltransferases and deacetylases, as well as protein kinases and phosphatases, have also been implicated in cocaine-induced behaviors (3, 6, 8, 11). More recently, regulation of the DNA methylation machinery [e.g., DNA methyltransferase 3A (Dnmt3A) and the methyl CpG binding protein 2] has been implicated in the actions of cocaine and other psychotstimulants, although the precise genomic loci affected by such adaptations remain unclear (12–14).
Although much work in recent years has focused on euchromatic chromatin remodeling in the development of addictive-like behaviors, very little attention has been placed on examining the potential consequences of repeated drug exposure on heterochromatic formation and genomic silencing in adult brain. One of the most heavily characterized markers of heterochromatin is trimethylated lysine 9 on H3 (H3K9me3). H3K9 can exist in a mono- (H3K9me1), di- (H3K9me2), or trimethylated state, in which multiple methyltransferase and demethylase enzymes act in concert to control distinct methylation profiles (15, 16). Di- and trimethylation of H3K9 create binding sites for chromodomain-containing proteins, including those of the heterochromatin protein 1 (HP1) family and are believed to promote transcriptional repression and genomic silencing through alterations in higher-order chromatin structure throughout euchromatic and constitutively heterochromatic genomic loci (17, 18). Although rare exceptions exist (e.g., ref. 19), the H3K9me3 mark, unlike H3K9me2 or H3K9me1, is thought to primarily reside in silenced, noncoding regions of the genome (20). Given its well-established role in mediating patterns of genomic repression, as well as the observation that its designated methylating enzymes (e.g., Suv39H1) exist in complexes with G9a (21) [previously implicated in cocaine action (9)], we hypothesized that H3K9me3 may play an important role in regulating cocaine-induced epigenomic landscapes in NAc.
Here, we report that both acute and repeated cocaine administration dynamically alters global levels of heterochromatic H3K9me3 in NAc. Reduced H3K9me3 expression after repeated, but not acute, cocaine persists during drug abstinence, indicating a putative role for heterochromatic regulation in the long-term actions of cocaine. We also use chromatin immunoprecipitation for H3K9me3 followed by massively parallel DNA sequencing (ChIP-Seq) to identify specific patterns of H3K9me3 regulation of the genome in response to repeated cocaine. The data suggest that chronic drug treatment causes the unsilencing of certain intergenic repetitive sequences, ultimately resulting in aberrantly expressed retrotransposable elements within the NAc.
Results
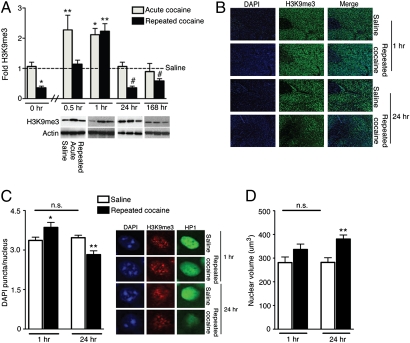
We first set out to identify whether H3K9me3 expression was globally influenced in NAc by several cocaine treatment regimens. Mice were treated with either acute (one dose) or repeated (seven daily doses) cocaine, and NAc samples, collected at various time points after cocaine administration, were analyzed for total levels of H3K9me3 by Western blotting. Surprisingly, although H3K9me3 is thought to be a highly stable heterochromatic mark (15), it displayed highly dynamic fluctuations in NAc in response to cocaine exposure (Fig. 1A). Acute cocaine rapidly increased H3K9me3 levels in NAc more than twofold above that of saline controls within 30 min, an effect that returned to control levels by 24 h. Animals treated repeatedly with cocaine, in contrast, displayed a delayed increase in H3K9me3 expression, taking approximately twice as long for H3K9me3 to reach levels comparable to those observed in acutely treated mice. This delayed methylation response occurred on top of a long-lasting suppression of this mark: H3K9me3 levels in the NAc were reduced by >50% at 24 h after the last dose of repeated cocaine, an effect that persisted for at least 1 wk of drug abstinence (Fig. 1A). Opposite regulation of H3K9me3 levels in NAc by repeated cocaine at 1 vs. 24 h was confirmed qualitatively by immunohistochemistry (Fig. 1B).
Fig. 1.
Cocaine dynamically regulates heterochromatin in NAc. (A) H3K9me3 levels are altered by cocaine administration. Mice were treated with saline or acute or repeated cocaine, and NAc samples were collected at 0.5, 1, 24, or 168 h (n = 5–8/group). Western blot analysis reveals a significant main effect (two-way ANOVA) of drug treatment (F2,68 = 6.439, P = 0.003), time after cessation of drug treatment (F3,68 = 11.21, P < 0.0001), and interaction between drug treatment and time (F6,68 = 3.891, P = 0.002) on H3K9me3 levels. All samples were normalized to actin, and data are represented as fold change from saline controls (–). Bonferroni post hoc tests indicate significant differences from saline controls (*P < 0.05, **P < 0.01) at 0.5 and 1 h. A posteriori Student t tests indicate significant differences from saline controls (#P < 0.05) at 24 and 168 h. The 0 and 24 h groups represent the same animals; 0 h is included to depict basal levels of H3K9me3 preceding cocaine administration for both the acute and repeated treatment groups. (B) Immunohistochemical analysis of mouse NAc qualitatively confirms increased H3K9me3 at 1 h and decreased H3K9me3 at 24 h after repeated cocaine administration. DAPI stains were used as a nuclear marker. All images were taken at 10× objective. (C) Altered heterochromatin domain numbers are consistent with altered levels of H3K9me3 after repeated cocaine at 1 and 24 h. Heterochromatin domains (puncta chromocenters) were quantified from DAPI-stained NAc sections at 1 h (n = 8/group) and 24 h (n = 10 to 11/group) after either saline or repeated cocaine. Approximately 112 H3K9me3/HP1+ neuronal nuclei were quantified from each animal at 40× objective. Statistical significance was determined using Student t tests (1 h-t16 = 2.159, *P < 0.05; 24 h-t21 = 3.730, **P < 0.01). Saline controls between 1 and 24 h were not significantly different (t19 = 0.6596, P > 0.05). (D) Repeated cocaine administration promotes increased nuclear volumes in NAc neurons after repeated drug administration. Nuclear volumes were quantified from DAPI+ nuclei obtained from NAc sections at 1 h (n = 6/group) and 24 h (n = 7–9/group) after saline or repeated cocaine. Approximately 20 nuclei were examined from each animal at 40× objective. Statistical significance was determined using Student t tests (1 h-t10 = 1.688, P > 0.05; 24 h-t14 = 3.610, **P < 0.01). Saline controls between 1 h and 24 h were not significantly different (t11 = 1.563, P > 0.05). All data are presented as average ± SEM.
As a control, we examined H3K9me3 levels in two other limbic nuclei implicated in cocaine action, the caudate putamen and medial prefrontal cortex, at 1 and 24 h after acute or repeated cocaine treatments. In contrast to the NAc, no alterations occurred in H3K9me3 levels in these brain regions (Fig. S1 A and B), indicating regional specificity to H3K9me3 regulation in NAc.
Dynamic yet persistent alterations in H3K9me3 expression in NAc after repeated cocaine exposure were surprising given the implication that cocaine might produce rapid swings in nuclear heterochromatin formation within the adult brain. To further study this possibility, we assessed other measures of heterochromatic repression in this brain region. Heterochromatin domains were identified in NAc nuclei by DAPI, a heterochromatin-binding fluorescent stain, and DAPI puncta were quantified at 1 and 24 h after repeated cocaine treatment. Only NAc neuronal nuclei were used for quantification, as determined by distinct nuclear morphological characteristics (22). Consistent with H3K9me3 levels, which were maximally increased at 1 h and maximally decreased at 24 h, we observed a significant increase in heterochromatin domains in NAc 1 h after repeated cocaine and a significant decrease at 24 h (Fig. 1C).
It has previously been shown that de-repression of chromatin can increase nuclear volume in cultured cells (23). This prompted us to examine whether altered levels of heterochromatin in NAc neuronal nuclei in response to repeated cocaine correlate, at a gross anatomical level, with alterations in nuclear volume. Nuclear volumes were calculated from DAPI-stained NAc nuclei at 1 and 24 h after repeated cocaine. At 24 h, we observed significantly enhanced nuclear volumes, consistent with the decrease in H3K9me3 levels seen at this time point (Fig. 1D). Because H3K9me3 is believed to be localized throughout noncoding, silenced regions of the genome, it is possible that repeated cocaine-induced reductions in H3K9me3 promote global patterns of genomic de-repression, ultimately resulting in chromatin de-condescension and nuclear expansion. In contrast, nuclear volume was not detectably altered at 1 h after repeated cocaine, which may simply reflect a lower limit of nuclear size for NAc neurons.
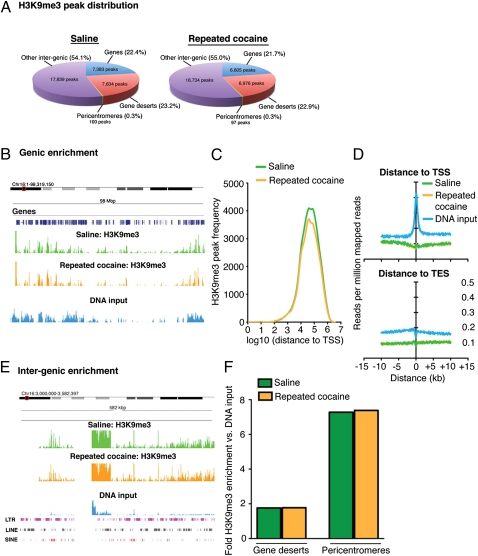
To more thoroughly investigate the genomic consequences of the sustained reduction in heterochromatic H3K9me3 in NAc after repeated cocaine, we used ChIP-Seq to identify specific genomic loci affected 24 h after the last drug dose. We identified 32,956 and 30,412 peaks in saline- and cocaine-treated animals, covering ≈420-Mbp and 383-Mbp genomic regions, respectively. Mapping saline- and cocaine-associated peaks to various genomic features (Methods) revealed that, under both saline and cocaine conditions, the large majority of H3K9me3 peaks reside in intergenic (i.e., noncoding) regions of the genome (Fig. 2A). These data are consistent with published reports of H3K9me3’s genomic localization in nonnervous tissue (24). Although fewer peaks were identified in NAc from cocaine-treated animals, this difference was not significant. This lack of difference in H3K9me3 peaks is in contrast to the decrease we observed in total H3K9me3 levels (Fig. 1A); however, two main considerations should be noted. First, to statistically identify genomic locations displaying regulation by cocaine, it must be assumed that alterations occurring at specific genomic loci reflect changes that are greater than any existing global phenomena; therefore, normalization methods all but assume that global changes do not exist. These normalization procedures are necessary to eliminate potential biases introduced during experimentation and may reflect an intrinsic limitation in the analysis of high-throughput ChIP-Seq data. Second, although total peak numbers did not differ significantly between saline and cocaine groups on a global scale, H3K9me3 enrichment differences within specific genomic regions (i.e., changes occurring on a much smaller scale in comparison with the entire genome) may account for fluctuations observed globally via Western blotting.
Fig. 2.
ChIP-Seq reveals significant enrichment of H3K9me3 throughout intergenic regions of the genome in NAc. (A) Mapping H3K9me3 peaks to several genomic features (Methods) reveals that the large majority of H3K9me3 peaks reside in intergenic genomic regions, whereas ≈20% associate with gene bodies. A small number of peaks reside within pericentromeres. No significant differences in peak distribution between saline- and cocaine-treated (24 h) animals were observed. (B) Although ≈20% of peaks reside within genic regions, H3K9me3 enrichment near gene bodies is low in comparison with DNA input. Image represents a Genome Browser snapshot of a highly genic 98-Mbp region of chromosome 16. (C) Analysis of H3K9me3 peak frequency in saline- and cocaine-treated animals demonstrates that the majority of peaks mapping to gene bodies are located more than 10 kbp from the TSS. Furthermore, H3K9me3 enrichment profiles for both treatment groups map below that of DNA input at both TSSs and TESs (D). (E) Consistent with peak distribution analyses, the H3K9me3 mark is heavily enriched throughout intergenic regions of the genome in comparison with DNA input. Image represents a 582-kbp region of the pericentromere of chromosome 16. Note the high abundance of genomic repeats overlapping with this region. (F) Although H3K9me3 is heavily enriched in noncoding regions of the genome, no significant differences were observed between saline and cocaine conditions within gene deserts or pericentromeres.
SICER-based peak distribution analysis indicated that 22.4% and 21.7% of peaks under saline and cocaine conditions, respectively, occurred near coding genes (Fig. 2A); however, enrichment profiles for H3K9me3 within these genic regions display only modest enrichment of the mark over DNA input. For example, Fig. 2B shows the lack of H3K9me3 genic enrichment on chromosome 16. This suggests a very limited role for H3K9me3 in regulating genomic repression in actively coding regions. Indeed, ChIP-Seq analyses of active and silent genes in NAc of saline and cocaine-treated animals revealed high levels of H3K9me3 enrichment exclusively within the bodies of silent genes, with a lack of H3K9me3 enrichment in active gene bodies (Fig. S2). Further data indicated that the H3K9me3 peaks mapping to gene bodies tend to be located more than 10 kbp away from transcription start sites (TSSs) (Fig. 2C). Additionally, binding intensity plots examining H3K9me3 reads around TSSs and transcription end sites (TESs) demonstrate that H3K9me3 is generally depleted at TSSs and TESs, with no differences observed between saline- and cocaine-treated samples (Fig. 2D). In contrast, as stated earlier, H3K9me3 peak distribution analyses indicated an abundance of this mark throughout intergenic regions of the genome (e.g., gene deserts, pericentromeres, etc.) (Fig. 2A), and enrichment profiles for these noncoding genomic regions further confirmed these findings; for example, the pericentromere of chromosome 16 displays high levels of H3K9me3 bound within this region in comparison with DNA input (Fig. 2E). No significant differences in H3K9me3 binding were observed globally between saline- and cocaine-treated animals along these intergenic regions (Fig. 2F).
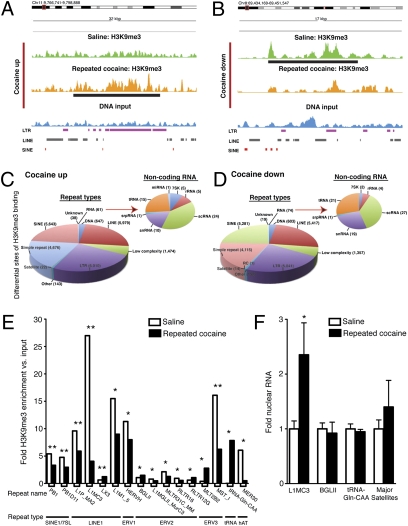
More detailed analysis of these intergenic regions, however, demonstrated many differential H3K9me3 enrichment sites in NAc between saline and cocaine conditions; examples of a cocaine-induced increase and decrease are shown in Fig. 3 A and B, respectively. Many of these sites correspond to regions containing a high abundance of genomic repeats, consistent with the knowledge that repetitive elements occur throughout intergenic regions of the genome. Therefore, we next assessed systematically the occurrence of differential H3K9me3 binding at repetitive elements genome-wide. More than 30,000 peak regions were evaluated for differences in read abundance; 1,644 and 1,365 cocaine up- and down-regulated regions were identified, respectively. More than 99% of these differential regions were shown to overlap with at least one repetitive element as defined by Repeat Masker (25).
Fig. 3.
Repeated cocaine administration alters H3K9me3 binding at repetitive elements and increases expression of retrotransposons in NAc. Representative Genome Browser images of repeated cocaine-induced (24 h) up- (A) and down- (B) regulated H3K9me3 enrichment throughout intergenic regions of the genome. Note that of the 1,644 and 1,365 cocaine up- vs. cocaine down-regulated regions, respectively, ≈99% of these regions overlap with at least one repetitive genomic sequences. Differential H3K9me3 binding site analyses after repeated cocaine treatment [cocaine up- (C) and cocaine down- (D) regulated regions] indicate altered H3K9me3 enrichment at numerous repeat types found in the genome, including LINE and SINE elements, LTRs, and noncoding RNAs. (E) Of the 973 genomic repeat elements assessed, as defined by RepBase (Methods), 17 repeats displayed significant changes in H3K9me3 enrichment after repeated cocaine treatment, with the large majority of these specific repeats displaying decreased H3K9me3 binding after repeated drug exposure (*P < 0.05, **P < 0.01). HERVK221 (2.3-fold decrease) was excluded because its fold change in comparison with DNA input is infinite. (F) qPCR analysis of nuclear RNA reveals significantly increased expression of L1MC3 (LINE-1 repeat) after repeated cocaine administration, with no changes observed in the expression of BGLII, tRNA-Gln-CAA, or Major Satellites between treatment groups. (LIMC3-t16 = 2.256, *P < 0.05; BGLII-t16 = 0.3459, P > 0.05; tRNA-Gln-CAA-t16 = 0.5273, P > 0.05; Major Satellites-t14 = 0.7824, P > 0.05) (n = 8 to 9/group).
A total of 24,693 repetitive elements [classified by repeat type, such as LINEs (long interspersed nuclear elements), SINEs (small interspersed nuclear elements), LTRs, etc.] displayed increased H3K9me3 binding after repeated cocaine (Fig. 3C), and 22,074 repetitive elements displayed decreased H3K9me3 binding (Fig. 3D). Although the majority of differential H3K9me3 binding sites were observed around genomic repeats, which tend to exist as constitutively silenced genomic DNA, a small percentage of changes were observed in genomic regions associated with noncoding RNAs (Fig. 3 C and D). Interestingly, after thorough examination of H3K9me3 binding abundance at 973 known repetitive elements, we observed that 17 displayed significant changes after repeated cocaine (Fig. 3E). These included the LINE-1 and SINE1/7S7 repeats, which represent distinct genomic retrotransposable elements. Retrotransposons are ubiquitous components of DNA and are defined as genetic elements that are capable of self-amplification within a genome. Consistent with our data that total H3K9me3 levels are reduced in NAc by repeated cocaine, we observed decreased H3K9me3 binding near several of these retrotransposons, with the most dramatic effects occurring near LINE-1 elements (e.g., L1MC3 as shown in Fig. 3E). While the majority of repeat types displayed decreased H3K9me3 binding, five displayed increased H3K9me3 association. Although unlikely, considering that most repetitive elements are constitutively silenced under basal conditions, it is possible that such increases in H3K9me3 enrichment after repeated cocaine may act to further repress specific genomic repeats. Interestingly, all five were H3K9me3 depleted under saline conditions.
To validate our observation of decreased H3K9me3 binding at, and genomic de-repression of, retrotransposons, NAc tissue was collected from separate cohorts of saline- and cocaine-treated mice and subjected to quantitative analyses of specific repeats. Quantitative PCR (qPCR) of nuclear RNA revealed a significant increase in levels of L1MC3 expression, a specific LINE-1 repeat, consistent with decreased heterochromatic silencing at this genomic locus (Fig. 3F). Interestingly, genomic repeats displaying increased H3K9me3 enrichment, such as BGLII and tRNA-Gln-CAA, as well as those that did not show significant H3K9me3 enrichment differences after cocaine (e.g., Major Satellites), did not display significant changes in nuclear expression (Fig. 3F).
Discussion
This study reveals the unique regulation by cocaine of heterochromatic histone methylation (H3K9me3) in the adult NAc and implicates heterochromatic mechanisms in long-term drug action. We show that both acute and repeated cocaine exposure promote dynamic fluctuations in total H3K9me3 levels in NAc, with repeated cocaine inducing persistent decreases in repressive methylation in this brain region. Decreased H3K9me3 expression after repeated cocaine correlates with reduced numbers of heterochromatin domains and increased nuclear volume, suggesting that a consequence of repeated cocaine is the de-repression of previously silenced chromatin and potential expansion of total genomic expression. Using ChIP-Seq, we show that H3K9me3 binding in NAc predominates in intergenic regions, and we demonstrate that repeated cocaine decreases H3K9me3 binding and unsilences several specific retrotransposons (e.g., LINE-1) in this brain region. Although the precise molecular mechanisms controlling H3K9me3 enrichment differences after cocaine remain unknown, these findings suggest a unique role for these ubiquitous mobile elements in cocaine action. It is also possible that cocaine exerts nonspecific or random effects on H3K9me3 enrichment throughout the genome, with little or inconsistent impact on neuronal function. Future experiments are needed to determine the genomic specificity of such changes, as well as the consequences of these events in the regulation of transcription and, ultimately, addiction-related behavior.
Our findings, along with previous demonstrations of the dynamic nature of total H3K9me3 levels in brain in response to other environmental perturbations (26), are surprising in light of current assumptions about this mark, which suggest that it exists as a highly stable regulator of heterochromatic repression, both on its own as well as via association with DNA methylation machinery (15, 27). Interestingly, although the levels of expression of specific chromatin-modifying enzymes responsible for this mark (e.g., Suv39H1) are not affected by cocaine exposure (9), numerous histone and DNA “writer/eraser” enzymes (e.g., G9a, SIRT1, Dnmt3a, etc.) and related proteins (21, 27, 28) that associate with H3K9me3 are regulated by the drug (5, 9, 12). These results suggest that cocaine-mediated alterations in H3K9me3 expression may be the result of disrupted chromatin remodeling complexes contributing to its deposition or maintenance. For example, G9a expression and activity are down-regulated by repeated cocaine, leading to decreased H3K9me2 levels in NAc (9). Although G9a is not believed to contribute directly to the establishment of the H3K9me3 mark (i.e., histone methylation likely does not occur in a processive manner) (16), G9a has been shown to exist in a complex with trimethylating enzymes for H3K9, such as Suv39H1 and Setdb1, with the functional activity of the complex dependent on the activity of its individual components (21). Therefore, it seems plausible that repeated cocaine-induced reductions in G9a expression may act to disrupt such complex formation, leading to persistently reduced levels of H3K9me3 in this brain region. Alternatively, such global changes in histone methylation after repeated cocaine may result from destabilization of the mark through altered regulation of histone demethylases, as well as through the establishment of neighboring histone modifications, which may directly affect the stability of H3K9me3 in structurally dense heterochromatic regions. Such possibilities highlight the potential for unique modes of H3K9me3 regulation in adult neurons, in comparison with the established biochemical modes of H3K9me3 regulation that occur during development.
Recent advances in ChIP-Seq technology have made it possible to achieve high-resolution genomic analyses of heterochromatic-associated histone marks, as well as the impact of cocaine treatment on such modifications. Although previous genome-wide promoter analyses of chromatin modifications in brain have proved very informative in terms of identifying unique patterns of gene regulation after cocaine treatment (5), we now demonstrate, using ChIP-Seq in an in vivo model of drug addiction, that repeated cocaine treatment also regulates histone modifications localized to heterochromatic regions of the genome. Consistent with previous literature (24), our data demonstrate the presence of H3K9me3 primarily throughout intergenic regions of the genome, such as gene deserts and pericentromeres, indicating a distribution of H3K9me3 in adult brain similar to that seen in other tissues. Together, our findings indicate that H3K9me3 mainly functions to direct heterochromatic, and not euchromatic, silencing, and therefore has very little effect on actively coding genes in brain.
In addition, using ChIP-Seq, we identify specific, localized patterns of differential H3K9me3 regulation in NAc after repeated cocaine treatment, in which ≈99% of regulated sites overlap with repetitive elements throughout these intergenic regions. Interestingly, upon further examination, we discovered that many of these changes in H3K9me3 binding occur in genomic repeats associated with retrotransposable elements, such as LINE-1s, SINEs, and LTRs. We subsequently validated these observations via qPCR, by demonstrating that some of these repeat sequences are indeed unsilenced, and show increased expression, in NAc in response to repeated cocaine. LINE-1 elements represent the most widespread classes of transposable elements found in the mammalian genome and account for ≈20% of total genomic DNA content (29). Although most LINE-1 elements are not considered to be actively transposable, ≈3,000 of these LINE-1s are believed to be potentially mobile within the mouse genome (30).
In recent years, much interest has arisen regarding the potential consequences of LINE-1 retrotransposition in the nervous system, after observations that LINE-1s are mobilized and genomically integrated during neurodevelopment and later in life during adult neurogenesis (31). Furthermore, using EGFP reporter mice allowing for visualization of LINE-1 insertion in adult neurons, voluntary exercise was found to increase LINE-1 genomic insertion specifically in neuronal progenitor cells in the subgranular zone of the hippocampus dentate gyrus (32). Although the functional consequences of LINE-1 mobilization and insertion in neurons remain unclear, it is conceivable that such integrations may contribute to alterations in the transcriptome through processes of transduction (i.e., exon shuffling, or insertion of unique transcriptional regulatory elements to new genomic loci), alternative splicing, transcript termination, or heterochromatization (31). Furthermore, it is not known whether such events are targeted or are simply random, as well as whether they occur within specific cells or represent more global phenomena within a particular circuit. However, surprisingly, LINE-1–mediated alterations in transcription after repeated cocaine offer a potentially unique mechanism through which chronic drug exposure might promote long-lasting changes in gene expression, contributing to the pathophysiology of drug addiction. Although many LINE-1 insertion events may be without consequence (i.e., many LINE-1 insertions presumably occur in intergenic regions such as gene deserts), some, if only a small percentage, could result in gene disruptions (loss or gain of function). Future studies are needed to identify whether (i) retrotransposon insertion events indeed affect transcriptional regulation of adult postmitotic neurons after cocaine exposure; (ii) such phenomena are targeted to specific genomic loci in a nonrandomized fashion, thereby allowing for direct examination of the genes being affected by cocaine-induced retrotransposition; (iii) mobilization occurs throughout multiple cell types or circuits or is targeted more specifically; and (iv) increased expression and mobilization of these elements actively contribute to cocaine-associated behavioral plasticity.
Together, our findings demonstrate a unique role for heterochromatin regulation in response to cocaine exposure and suggest that a potential consequence of chronic cocaine-mediated heterochromatic de-repression is the unsilencing of retrotransposable elements in NAc neurons. These events may ultimately act to disrupt the transcriptome of distinct neuronal populations, leading to altered behavioral sensitivity to drugs of abuse. Identifying how these mechanisms contribute to drug-induced behaviors will further aid in our understanding of the molecular mechanisms controlling cocaine addiction and may open doors for the future development of novel therapeutics aimed at alleviating this devastating condition.
Methods
Animals and Western Blots.
Detailed methods are provided in SI Methods.
Immunohistochemistry.
Mice were sedated with chloral hydrate and perfused with 4% paraformaldehyde before being analyzed by single or double immunohistochemistry, as previously described (9). See SI Methods for details.
ChIP-Seq.
Chromatin immunoprecipitation and sequencing preparation.
ChIP was performed using previously validated methods (9, 33, 34). See SI Methods for detailed ChIP and sequencing preparation methods.
Peak finding.
Two and three ChIP-Seq runs for NAc tissue obtained from cocaine- and saline-treated mice, respectively, were obtained from Illumina’s GAII sequencing machine. Short reads of 36 bp were first analyzed by Illumina’s pipeline software for quality filtering and aligned to the reference genome. Only uniquely aligned reads were kept for further analysis. To remove potential PCR amplification and sequencing bias, only two copies of the same strand read that successfully mapped to a genomic location were preserved. This level of redundancy (up to two copies) was determined by assuming a uniform binomial distribution across the genome (35). Sequencing runs were then pooled under each condition to provide better coverage than could be obtained within a single run. Two ChIP-Seq libraries were achieved with more than 19 million and 30 million reads for cocaine and saline, respectively. Per our terminology, we use “run” to indicate the data of a separate ChIP-Seq sample and “library” to indicate the pooled data of ChIP-Seq samples under a specific condition. The SICER peak-finding algorithm (36) was used to identify the H3K9me3-enriched sites throughout the genome. SICER is specifically designed to meet the needs for peak finding with histone marks that have long, diffusive patterns of enrichment. SICER was run on each ChIP-Seq library, along with a DNA input library, of more than 38 million monoclonal reads. We used a DNA input library to eliminate biases introduced by sequencing, chromatin structure, and sequence compositions, and thereby better control for false positives. In SICER's parameter settings, we used the default false discovery rate (FDR) of 1e-3 and set the window size to be 1,000 bp. The gap size was determined as suggested in (36) (Fig. S3).
Peak mapping to genomic features.
The TSSs and TSEs of all Refseq genes were determined by the refFlat.txt program downloaded from the University of California, Santa Cruz genome browser (25), with more than 26,000 pairs extracted. A gene body was determined as the genomic interval from 2 kbp upstream of the TSS to 500 bp downstream of the TES so that proximal regulatory regions were also included. A peak was assigned to a Refseq gene if the center of the peak was within the gene body. Gene deserts and pericentromeres were determined using the same approach as described in ref. 24. Each gene desert must have represented a region greater than 1 Mbp between two adjacent genes, with 10 kbp proximal to each gene removed. We therefore defined 299 gene deserts on the mouse genome mm9. Pericentromeres were defined for each chromosome as the region between the two nearest genes to the centromeres, and the 10 kbp proximal to the genes were removed. A peak was assigned to either a gene desert or a pericentromere if it overlapped with the region as defined above. All remaining peaks that were not assigned to any of the above regions were considered to reside within other intergenic regions.
Calculating binding enrichment in intergenic regions.
We used the same approach as described elsewhere (24) to determine the enrichment of H3K9me3 in gene deserts and pericentromeres. The number of ChIP-Seq reads was compared with that of the DNA input reads mapped to each region. To adjust for the different library sizes, we divided the number of reads by its corresponding library size, so that the percentage of mapped reads was actually being compared. Assuming that we doubled the size of a library, we would expect that the number of reads mapped to each region would be doubled as well. However, the percentage of the reads mapped to that specific region should theoretically stay the same.
Determining read abundance in repetitive elements.
Each repetitive element is represented repeatedly throughout the genome, thus quantification of sequence abundance in each of these regions is difficult because most sequence aligners will drop reads that are mapped ambiguously. To overcome this obstacle, we first downloaded the consensus sequences of 973 mouse repetitive elements from Repbase (37) and used them to construct a pseudo “repetitive genome.” As a metaphor, each repetitive element can be viewed as a stand-alone chromosome in the repetitive genome. We then used the BWA sequence aligner (38) to map each ChIP-Seq run to the repetitive genome and calculate the numbers of reads mapped.
To determine the statistical significance of the differences between saline and cocaine conditions, we accounted for biological variability using DESEq (39). DESeq uses a negative binomial model to approximate the null distribution assuming that the two conditions exhibit the same read abundance. It uses an empirical Bayesian approach to estimate the variance among biological replicates by borrowing neighborhood information. The empirical Bayesian method allows DESeq to provide more reliable estimations of the variance than if we were to use single regions alone in the case of small numbers of replicates. It therefore avoids overoptimistic estimation of P values and better controls for false positives. We finally identified repetitive elements displaying different read abundances between the two conditions based on P value <0.05 and a fold change ≥1.3. The fold enrichment of the H3K9me3 in repetitive elements against DNA input was then determined using the same procedure as in gene deserts and pericentromeres.
Identifying differential regions by read abundance.
To obtain a detailed map of differential H3K9me3 binding sites throughout the genome, we used the two peak lists described above as a filter to focus on regions that display potential binding events. This greatly reduces the chance that signals are being buried by noise. The numbers of reads mapped to those peak regions were then determined for each biological replicate under the two conditions, and the significance of differences was assessed using DESeq, as described above. Each peak list was used to define regions showing enrichment under the corresponding condition, so that inconsistencies between the two peak lists were avoided. A cutoff P value of <0.05 and fold change ≥1.3 was used to identify differential regions between saline and cocaine conditions.
Nuclear RNA Isolation and qPCR.
Nuclear extraction (40) and qPCR (9) were performed as previously described. See SI Methods for detailed methods and primer sequences (Table S1).
Statistics.
See SI Methods for detailed statistical descriptions.
Acknowledgments
We thank John Greally (Albert Einstein College of Medicine) for helpful discussions. This work was supported by grants from the National Institute on Drug Abuse.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. GSE24850).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1015483108/-/DCSupplemental.
References
- 1.Hyman SE, Malenka RC, Nestler EJ. Neural mechanisms of addiction: the role of reward-related learning and memory. Annu Rev Neurosci. 2006;29:565–598. doi: 10.1146/annurev.neuro.29.051605.113009. [DOI] [PubMed] [Google Scholar]
- 2.Kumar A, et al. Chromatin remodeling is a key mechanism underlying cocaine-induced plasticity in striatum. Neuron. 2005;48:303–314. doi: 10.1016/j.neuron.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 3.Renthal W, et al. Histone deacetylase 5 epigenetically controls behavioral adaptations to chronic emotional stimuli. Neuron. 2007;56:517–529. doi: 10.1016/j.neuron.2007.09.032. [DOI] [PubMed] [Google Scholar]
- 4.Renthal W, et al. Delta FosB mediates epigenetic desensitization of the c-fos gene after chronic amphetamine exposure. J Neurosci. 2008;28:7344–7349. doi: 10.1523/JNEUROSCI.1043-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Renthal W, et al. Genome-wide analysis of chromatin regulation by cocaine reveals a role for sirtuins. Neuron. 2009;62:335–348. doi: 10.1016/j.neuron.2009.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stipanovich A, et al. A phosphatase cascade by which rewarding stimuli control nucleosomal response. Nature. 2008;453:879–884. doi: 10.1038/nature06994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wang L, et al. Chronic cocaine-induced H3 acetylation and transcriptional activation of CaMKIIalpha in the nucleus accumbens is critical for motivation for drug reinforcement. Neuropsychopharmacology. 2010;35:913–928. doi: 10.1038/npp.2009.193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brami-Cherrier K, et al. Parsing molecular and behavioral effects of cocaine in mitogen- and stress-activated protein kinase-1-deficient mice. J Neurosci. 2005;25:11444–11454. doi: 10.1523/JNEUROSCI.1711-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maze I, et al. Essential role of the histone methyltransferase G9a in cocaine-induced plasticity. Science. 2010;327:213–216. doi: 10.1126/science.1179438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 11.Levine AA, et al. CREB-binding protein controls response to cocaine by acetylating histones at the fosB promoter in the mouse striatum. Proc Natl Acad Sci USA. 2005;102:19186–19191. doi: 10.1073/pnas.0509735102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.LaPlant Q, et al. Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci. 2010;13:1137–1143. doi: 10.1038/nn.2619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Im HI, Hollander JA, Bali P, Kenny PJ. MeCP2 controls BDNF expression and cocaine intake through homeostatic interactions with microRNA-212. Nat Neurosci. 2010;13:1120–1127. doi: 10.1038/nn.2615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Deng JV, et al. MeCP2 in the nucleus accumbens contributes to neural and behavioral responses to psychostimulants. Nat Neurosci. 2010;13:1128–1136. doi: 10.1038/nn.2614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Peters AH, et al. Partitioning and plasticity of repressive histone methylation states in mammalian chromatin. Mol Cell. 2003;12:1577–1589. doi: 10.1016/s1097-2765(03)00477-5. [DOI] [PubMed] [Google Scholar]
- 16.Rice JC, et al. Histone methyltransferases direct different degrees of methylation to define distinct chromatin domains. Mol Cell. 2003;12:1591–1598. doi: 10.1016/s1097-2765(03)00479-9. [DOI] [PubMed] [Google Scholar]
- 17.Bannister AJ, et al. Selective recognition of methylated lysine 9 on histone H3 by the HP1 chromo domain. Nature. 2001;410:120–124. doi: 10.1038/35065138. [DOI] [PubMed] [Google Scholar]
- 18.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 19.Muller-Tidow C, et al. Profiling of histone H3 lysine 9 trimethylation levels predicts transcription factor activity and survival in acute myeloid leukemia. Blood. 2010;116:3564–3571. doi: 10.1182/blood-2009-09-240978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Barski A, et al. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 21.Fritsch L, et al. A subset of the histone H3 lysine 9 methyltransferases Suv39h1, G9a, GLP, and SETDB1 participate in a multimeric complex. Mol Cell. 2010;37:46–56. doi: 10.1016/j.molcel.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 22.Matamales M, et al. Striatal medium-sized spiny neurons: Identification by nuclear staining and study of neuronal subpopulations in BAC transgenic mice. PLoS ONE. 2009;4:e4770. doi: 10.1371/journal.pone.0004770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bártová E, et al. Nuclear levels and patterns of histone H3 modification and HP1 proteins after inhibition of histone deacetylases. J Cell Sci. 2005;118:5035–5046. doi: 10.1242/jcs.02621. [DOI] [PubMed] [Google Scholar]
- 24.Rosenfeld JA, et al. Determination of enriched histone modifications in non-genic portions of the human genome. BMC Genomics. 2009;10:143. doi: 10.1186/1471-2164-10-143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kent WJ, et al. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hunter RG, McCarthy KJ, Milne TA, Pfaff DW, McEwen BS. Regulation of hippocampal H3 histone methylation by acute and chronic stress. Proc Natl Acad Sci USA. 2009;106:20912–20917. doi: 10.1073/pnas.0911143106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Henckel A, et al. Histone methylation is mechanistically linked to DNA methylation at imprinting control regions in mammals. Hum Mol Genet. 2009;18:3375–3383. doi: 10.1093/hmg/ddp277. [DOI] [PubMed] [Google Scholar]
- 28.Vaquero A, et al. SIRT1 regulates the histone methyl-transferase SUV39H1 during heterochromatin formation. Nature. 2007;450:440–444. doi: 10.1038/nature06268. [DOI] [PubMed] [Google Scholar]
- 29.Lander ES, et al. International Human Genome Sequencing Consortium Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 30.Goodier JL, Ostertag EM, Du K, Kazazian HH., Jr A novel active L1 retrotransposon subfamily in the mouse. Genome Res. 2001;11:1677–1685. doi: 10.1101/gr.198301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Singer T, McConnell MJ, Marchetto MC, Coufal NG, Gage FH. LINE-1 retrotransposons: Mediators of somatic variation in neuronal genomes? Trends Neurosci. 2010;33:345–354. doi: 10.1016/j.tins.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Muotri AR, Zhao C, Marchetto MC, Gage FH. Environmental influence on L1 retrotransposons in the adult hippocampus. Hippocampus. 2009;19:1002–1007. doi: 10.1002/hipo.20564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lee TI, Johnstone SE, Young RA. Chromatin immunoprecipitation and microarray-based analysis of protein location. Nat Protoc. 2006;1:729–748. doi: 10.1038/nprot.2006.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilkinson MB, et al. Imipramine treatment and resiliency exhibit similar chromatin regulation in the mouse nucleus accumbens in depression models. J Neurosci. 2009;29:7820–7832. doi: 10.1523/JNEUROSCI.0932-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang Y, et al. Model-based analysis of ChIP-Seq (MACS) Genome Biol. 2008;9:R137. doi: 10.1186/gb-2008-9-9-r137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zang C, et al. A clustering approach for identification of enriched domains from histone modification ChIP-Seq data. Bioinformatics. 2009;25:1952–1958. doi: 10.1093/bioinformatics/btp340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jurka J, et al. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 38.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ogita K, et al. Localization of activator protein-1 complex with DNA binding activity in mitochondria of murine brain after in vivo treatment with kainate. J Neurosci. 2002;22:2561–2570. doi: 10.1523/JNEUROSCI.22-07-02561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]