Abstract
Spinocerebellar ataxia 10 (SCA10) is an autosomal dominant disease caused by large-scale expansions of the (ATTCT)n repeat within an intron of the human ATXN10 gene. In contrast to other expandable repeats, this pentanucleotide repeat does not form stable intra- or interstranded DNA structures, being a DNA unwinding element instead. We analyzed the instability of the (ATTCT)n repeat in a yeast experimental system, where its expansions led to inactivation of the URA3 reporter gene. The inactivation was due to a dramatic decrease in the mRNA levels owing to premature transcription termination and RNA polyadenylation at the repeat. The rates of expansions strongly increased with the repeat's length, mimicking genetic anticipation in human pedigrees. A first round of genetic analysis showed that a functional TOF1 gene precludes, whereas a functional RAD5 gene promotes, expansions of the (ATTCT)n repeat. We hypothesize that repeat expansions could occur upon fortuitous template switching during DNA replication. The rate of repeat contractions was elevated in the Tof1 knockout strain, but it was not affected by the RAD5 gene. Supporting the notion of replication irregularities, we found that (ATTCT)n repeats also cause length-dependent chromosomal fragility in yeast. Repeat-mediated fragility was also affected by the Tof1 and Rad5 proteins, being reduced in their absence.
Keywords: DNA repeats, genome instability, DNA repair
Expansion of simple DNA repeats is a cause of more than 30 hereditary diseases in humans (1). Just one repeat within a particular gene undergoes expansions in each case, suggesting that expansion events occur in cis, in contrast with trans-acting mutations of DNA metabolism destabilizing different repeats (2–4). Many of the repeat expansion diseases are characterized by genetic anticipation (i.e., an increased severity and early onset of the disease as the repeat progressively expands during intergenerational transmissions) (5). The exact mechanisms of repeat expansions in humans are unknown, although data from model systems implicate DNA replication, repair, and recombination as contributors to repeat expansions (6).
Trinucleotide, tetra-, penta-, and dodecanucleotide repeats can expand, leading to disease (1). It was generally believed that unusual secondary structures formed by expandable repeats are central for the expansion process (7). Recently, however, it has become clear that this is not always the case. A pentanucleotide repeat, (ATTCT)n, large-scale expansions of which cause spinocerebellar ataxia type 10 (SCA10) (8), does not form any unusual structures (9) but is a DNA unwinding element (DUE). Because a DUE is involved in replication initiation in vivo, it was proposed that (ATTCT)n repeats expansions could result from multiple reinitiation of DNA replication within this sequence (9, 10). Another example of an expandable repeat that is not expected to form alternative DNA structures is a sequence, (TGGAA)n, responsible for SCA31 (11). We were interested, therefore, in whether the same mechanisms govern expansions of the structure-prone and structure-proof repeats.
Here, we concentrated on the mechanisms of expansions of the SCA10 (ATTCT)n repeat. This repeat is positioned in the ninth intron of the human ataxin-10 (ATXN10) gene (8). Normal individuals have 10–22 copies, whereas affected individuals may have up to 4,500 copies of this repeat. SCA10 is an autosomal dominant disease that is prevalent in Mexico and Brazil. The two populations differ both symptomatically and genetically when it comes to the disease (12). In addition to ataxia, Mexican families are afflicted with epilepsy, but Brazilian families are not (13, 14). At a DNA level, expanded (ATTCT)n repeats without interruptions are typical for the Mexican population, whereas multiple interruptions throughout the expanded repetitive run are characteristic for the Brazilian population. The SCA10 pathogenesis is not well understood. It was suggested that haploinsufficiency of the ATXN10 gene, RNA gain of function (15), or chromatin change (12, 16) could contribute to the disease.
The mechanisms responsible for the expansions of the (ATTCT)n repeats remain unclear. They were never detected in any experimental system, making the genetic analysis of the process impossible. Thus, from a biomedical point of view it was paramount to develop a genetically tractable experimental system to study (ATTCT)n repeat expansions. Here we achieved this goal using our recently developed strategy to monitor large-scale changes in repeat lengths in yeast (17). Specifically, noninterrupted ATTCT repeats, ranging from 46 to 81 copies, were integrated into an artificial intron of the URA3 gene, which rendered the gene functional. Expansions of the repeat beyond ≈85 copies blocked the reporter gene's expression, leading to the selectable genotype, 5-fluoroorotic acid resistance. The rate of repeat expansions seemed to increase with the repeat's length, mimicking human pedigrees. We identified several proteins that affected repeat expansions. The Tof1 protein, a component of the replication fork stabilizing complex (18, 19), seemed to prevent (ATTCT)n repeat expansions, whereas the Rad5 protein, responsible for template switching during postreplication repair (20), was necessary for repeat expansions. At the same time, the Rad52 protein, a master component of homologous recombination in yeast (21), does not seem to play a significant role in (ATTCT)n repeat expansions. We also found that (ATTCT)n repeats stimulated chromosomal fragility in a length-dependant manner and that fragility was also affected by the Tof1 and Rad5 proteins, being reduced in their absence.
We conclude that repeat expansions occur during DNA replication and/or postreplicative repair. Remarkably, our genetic data are qualitatively similar to that obtained for a different repeat, (GAA)n—a prominent structure-forming repeat. We conclude, therefore, that the repetitive nature of these sequences might be the key factor that predisposes them to expansions during DNA replication and propose a model for this process. This said, comparison of expansion and contraction rates for different repeats confirms that the structure-forming potential of a sequence dramatically contributes to its instability.
Results
Yeast System to Study Expansions of (ATTCT)n Repeats.
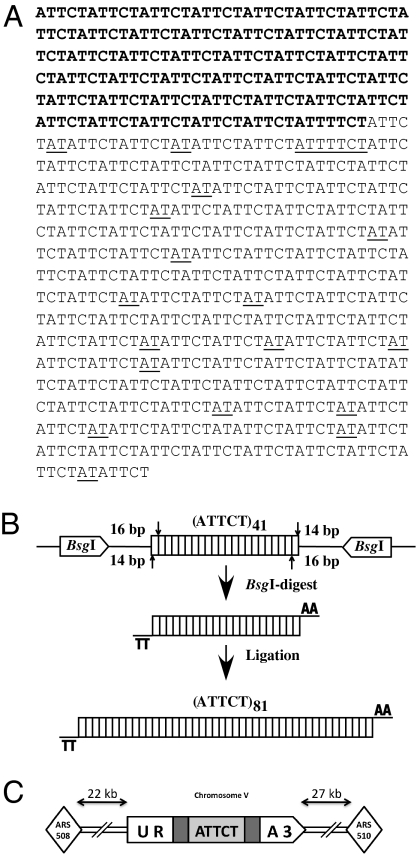
A DNA fragment from a patient with SCA10 containing more than 500 (ATTCT)n repeats was initially cloned into the pcDNA3.1/Hygro vector as previously described (15). After we transformed the resultant plasmid into Escherichia coli, the repeat had contracted down to ≈220 units. Sequencing of the contracted repeat revealed multiple interruptions providing only 41 ATTCT repeats as the longest uninterrupted run of repeats (Fig. 1A). We amplified this noninterrupted run using PCR with the primers carrying restriction sites for the enzyme BsgI at their 5′ ends. Upon digesting a PCR product, an (ATTCT)41•(AGAAT)41 duplex with AA and TT 3′ overhangs was formed (Fig. 1B). These overhangs allow a self-ligation of multiple (ATTCT)41•(AGAAT)41 fragments in a head-to-tail orientation only. Using this approach, we generated an uninterrupted (ATTCT)81 repeat in vitro.
Fig. 1.
Yeast system for studying expansions of (ATTCT)n repeats. (A) An (ATTCT)n repeat from a Brazilian patient with SCA10 contains multiple interruptions (underlined), providing only 41 uninterrupted repeats (bolded). (B) Scheme for cloning longer noninterrupted (ATTCT)n repeats. (C) Our experimental cassette, in which various (ATTCT)n repeats (light gray) are placed within an intron (dark gray) of the artificially split URA3 gene (white) on chromosome V. This cassette replaced the endogenous URA3 gene, positioned 22 kb away from ARS508 on the left and 27 kb away from ARS510 on the right.
The (ATTCT)81 repeat was then cloned into the intron of the artificially split URA3 gene (17). The resultant cassettes were excised and integrated into chromosome V of the CH1585 strain, replacing its ura3-52 allele upon selection for uracil prototrophy (Fig. 1C). PCR analysis of the URA+ clones revealed properly integrated URA3 cassettes with 46, 64, and 81 ATTCT repeats, which corresponded to 628-, 718-, and 803-bp-long introns. A yeast strain with 81 ATTCT repeats in the URA3 gene grew exceptionally slowly on the media lacking uracil. Furthermore, when it was plated on the 5-FOA–containing media, a lawn of small colonies would slowly form as well. We concluded, therefore, that 81 ATTCT repeats inactivated the URA3 gene strongly enough to make cells partially 5-FOA resistant, which made this repeat useless for further selection. This left us with two repeat lengths suitable for the selection for expansions: (ATTCT)46 and (ATTCT)64.
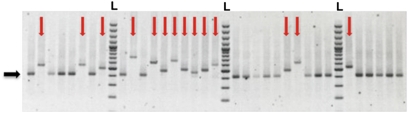
In a previous study of (GAA)n repeats in yeast, we found that repeat expansions blocked splicing of the URA3 gene carrying an intron, when the intron's length exceeded ≈1.1 kb (17). We reasoned that substantial expansions of the (ATTCT)n repeats within the URA3 intron should lead to the reporter's inactivation as well, making yeast 5-FOA resistant. Fig. 2 shows PCR analysis of the repeat lengths 5-FOAR clones originated in the strain carrying 64 (ATTCT)n repeats in the URA3 intron. Two types of events are evident: significant repeat expansions and unchanged repeat lengths.
Fig. 2.
PCR analysis of 5-FOA–resistant clones. The characteristic results for 5-FOAR clones originated from (ATTCT)64 repeats. Red vertical arrows point to expanded repeats, whereas the black horizontal arrow marks the position of the original repeat. L, 100 bp-plus ladder (Fermentas).
Rates and Scales of (ATTCT)n Repeat Expansions.
To determine the rates of both events leading to drug resistance, 8–12 independent single colonies grown on full media were replated onto the selective, 5-FOA–containing media, as well as on full media for normalization. All 5-FOAR clones from six to eight selective plates were analyzed by PCR for their repeat length. This gave us the frequencies of expansions of both events for (ATTCT)n repeats. Their rates were then calculated using the method of mutant accumulation, as previously described (17). The average rate of expansions increased eightfold when the number of repeats increased 1.4-fold (Table 1). The difference in expansion rates between (ATTCT)46 and (ATTCT)64 repeats was highly statistically significant (P < 0.0001). The rates of events in which repeat lengths remained unchanged also depended on the repeat's length, but very modestly: a 2.3-fold increase when the number of repeats increased 1.4-fold. Surprisingly, only three of 16 sequenced clones in this group contained mutations in the URA3 cassette: two had missense mutations in the URA3 ORF, and one had a point substitution in the ACT1 intron. The remaining 81% of 5-FOA–resistant clones did not have any mutations in the URA3 cassette (discussed in SI Results).
Table 1.
Rates (95% confidence intervals) of expansions and other events leading to 5-FOA resistance
| Repeat number | Rate of expansions (×10−7) | Rate of unchanged repeats (×10−7) | Rate of mutations (×10−7) |
| 0 | N/A | N/A | 0.94 (0.24–2.3) |
| 46 | 0.44 (0.35–0.79) | 3.5 (2.4–4.6) | Not studied |
| 64 | 3.3 (2.4–4.1) | 7.6 (5.6–9.7) | 1.4 (1.0–1.8) |
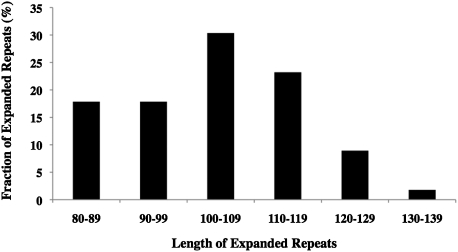
Because the expansion rate for the (ATTCT)64 repeat was sufficiently high (≈4 × 10−7 per cell/per generation), we accumulated 56 independent clones to built a distribution for the lengths of expanded repeats (Fig. 3). From this distribution, the range of expansion lengths lies between 81 and 132 repeats (i.e., the expansions are fairly significant). Evidently, the distribution is biased owing to the selection cutoff around 80–85 repeats. This is consistent with previous data that the URA3 cassette with the (ATTCT)81 repeat makes yeast partially 5-FOA resistant. Importantly, however, the median length of expanded repeats corresponds to ≈105, which is above and beyond the selection cutoff. We believe that an addition of 40 pentanucleotide repeats (200 bp) might reflect an average increment in the expansions of (ATTCT)n repeats.
Fig. 3.
Length distribution among the expanded repeats originated from the original (ATTCT)64 repeat. The range is from 81 to 132 repeats, the mean expansion corresponds to 104 repeats, and the median expansion length is 105 repeats.
Effects of (ATTCT)n Repeats on Reporter Gene Expression.
A priori, repeat-containing clones could become drug resistant if one of the following events occurred: (i) repeats expanded to a point where the intron length exceeded 1.1 kb (22), or (ii) expanded repeats directly block expression of the URA3 gene at the transcription or posttranscription level. The latter scenario seemed more likely for (ATTCT)n repeats, because a strain carrying 81 repeats creates an 803-bp-long intron of the URA3 cassette and is partially 5-FOA resistant.
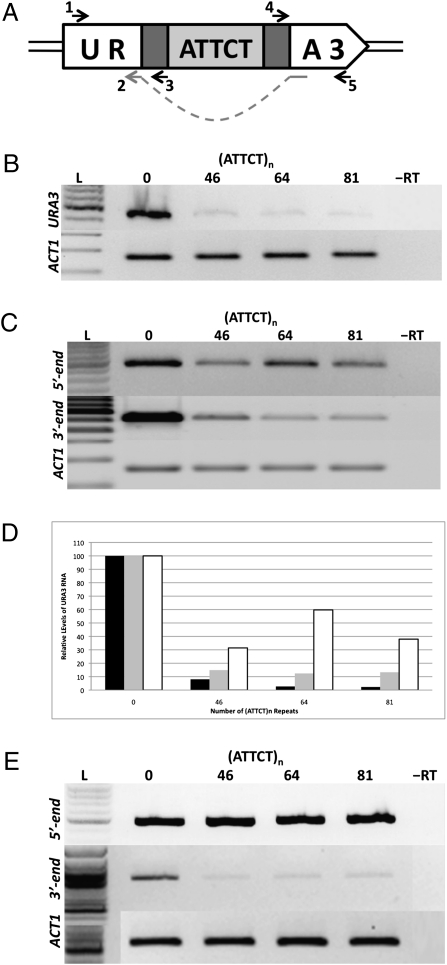
To study the effects of the (ATTCT)n repeats on the expression of the URA3 gene, we first analyzed the levels of its mRNA by RT-PCR. We first conducted reverse transcription reaction with random primers followed by PCR with mRNA-specific primers (Fig. 4A). One can see that even the shortest repeat studied, (ATTCT)46, decreased the amount of URA3 mRNA by 10-fold compared with the URA3 cassette with no repeats. As the numbers of repeats increased, there were progressively lesser amounts of URA3 mRNA (Fig. 4B). With the longest repeat, (ATTCT)81, less than 2% of the amount for the control mRNA was detected. This very low level of expression of the URA3 gene, carrying 81 (ATTCT)n repeats, explains the ability of the corresponding yeast strain to slowly grow on both URA– media and 5-FOA+ media. Note that a 10-fold decrease in the URA3 expressions does not make the yeast strain a uracil auxotroph, because orotidine-5′-phosphate decarboxylase is an exceptionally proficient enzyme (23).
Fig. 4.
Effects of (ATTCT)n repeats on the reporter's gene expression. (A) Schematic representation of the repeat-bearing URA3 cassette together with various primers used for the RT-PCR. Because primer 2 contains exonic sequences surrounding the intron, primers 1 and 2 were used to specifically amplify URA3 mRNA. Primers 3 and 4 contain intronic sequences. Consequently, primers 1 and 3 were used to amplify URA3 pre-mRNA upstream of the repeat (5′ end), whereas primers 4 and 5 amplified pre-mRNA downstream of the repeat (3′ end). (B) RT-PCR analysis of URA3 mRNA for clones containing 0, 46, 64, and 81 ATTCT repeats. Actin mRNA was used for the normalization. (C) RT-PCR analysis of URA3 pre-mRNA upstream and downstream of the repeat tract. (D) Quantitative graph showing relative amounts of URA3 mRNA and pre-mRNA. Black, gray, and white bars correspond to mRNA, pre-mRNA downstream of the repeat, and pre-mRNA upstream of the repeat, respectively. (E) RT-PCR analysis of the URA3 RNA upstream and downstream of the repeat tract for the polyadenylated transcripts only.
Further RT-PCR experiments were performed to examine the levels of the URA3 pre-mRNA situated 5′ and 3′ to the (ATTCT)n repeats. The levels of pre-mRNA downstream of the repeats decreased strongly with an increase in their lengths (Fig. 4C). In contrast, the levels of pre-mRNA upstream of the repeats were only modestly decreased compared with the repeat-free control (Fig. 4C).
Two scenarios could lead to a dramatic difference in pre-mRNA level upstream and downstream of the (ATTCT)n repeat: (i) transcription could be stalled by the repetitive sequence per se, or a protein bound to it, or (ii) (AUUCU)n runs in the transcript could set off RNA polyadenylation leading to premature transcription termination. To distinguish between these possibilities, we modified RT-PCR to amplify only polyadenylated transcripts by using oligo(dT) primers for the reverse transcription reaction. Fig. 4E shows that equal amounts of URA3 pre-mRNA upstream of the repeat are present for all repeat lengths in this setting. This strongly suggests that the repetitive tract signals unruly polyadenylation.
Test of (ATTCT)n Repeat Fragility.
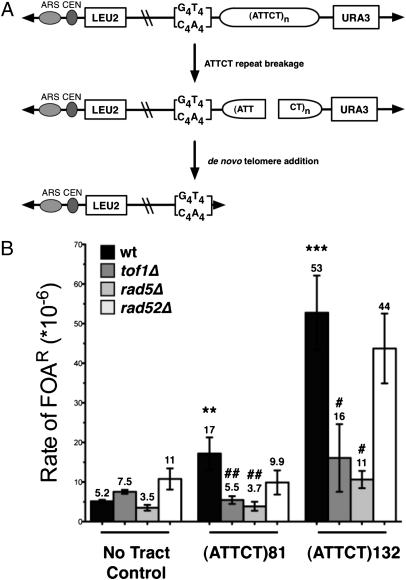
Expanded trinucleotide repeats, as well as some AT-rich minisatellites, are sites of increased chromosomal fragility in human and yeast cells (24). To test whether the expanded SCA10 repeat would be a fragile site, we used a previously designed assay to assess the breakage rate of a yeast artificial chromosome (YAC) containing an expanded (ATTCT)n repeat. In this assay (Fig. 5A), loss of the right arm of the YAC distal to the repeat tract can be measured by loss of the URA3 marker and subsequent ability of the cells to grow on 5-FOA media (25). Only breakage events that fail to heal normally and instead result in arm loss are measured, thus the rate of 5-FOA resistance is an underestimation of the true breakage rate, but it can be used to obtain a relative rate compared with a no-tract control, in this case a 386-bp nonrepetitive AT-rich human sequence. When we initially cloned the (ATTCT)81 repeat into the YAC, we also obtained a spontaneous expansion in one of the transformants, which was verified to be a pure (ATTCT)132 tract by sequencing. The (ATTCT)81 tract induced 3.3-fold more chromosome fragility than the control sequence of equal length (P = 0.003), and the longer (ATTCT)132 tract led to a 10.3-fold increase, a highly significant difference compared with the no-tract control (P < 0.0001) (Fig. 5B). Thus, expanded SCA10 repeats induce fragility in a length-dependent manner. The level of fragility observed here for the (ATTCT)n repeats seems to be comparable to that of a (CAG)n repeat tested in the same assay (25–27).
Fig. 5.
Effect of (ATTCT)81 and (ATTCT)132 repeats on chromosome fragility. (A) Diagram of the fragility assay. The (ATTCT)n repeat tracts were cloned near the end of a YAC, proximal to a URA3 marker gene. Chromosomes that break at or near the repeat and fail to repair will lose the URA3 gene and can be rescued by telomere addition onto the G4T4 telomere seed sequence. (B) Rate of Leu+FOAR cells provides a relative breakage rate compared with a no-tract control. Significance was determined by a pooled variant t test: compared with the no tract control, *P < 0.05, **P < 0.01, ***P < 0.001; or compared with the same tract length in the wild-type (wt), #P < 0.05, ##P < 0.01.
Genetic Analysis of Repeat Expansions, Contractions, and Fragility.
To gain more insight into the mechanisms of repeat expansions, we analyzed the effects of various mutations affecting DNA replication, recombination, and repair on this process. Specifically, we analyzed three mutations: Δtof1, Δrad5, and Δrad52, because they were useful for our understanding of the (GAA)n repeat expansions (17).
A URA3 cassette containing (ATTCT)64 repeats was introduced in Tof1, Rad5, or Rad52 knockout strains, and the rates of the repeat expansions in those strains were determined. The median rates expansions obtained from at least three independent experiments are presented in Table 2. One can see that in the tof1Δ strain, the average rate of expansions was fivefold higher than that in the wild-type train (P < 0.0001), suggesting that Tof1p works against repeat expansions. The results in rad5Δ strain were strikingly opposite: no expansions at all were observed, indicating that the Rad5 protein could be the key player promoting repeat expansions. The rate of expansions in rad52Δ was threefold less than in the wild-type strain, a difference that was not as dramatic as in the rad5Δ strain but still statistically significant.
Table 2.
Genetic control of expansions and contractions of the (ATTCT)n repeats
| Genetic background | (ATTCT)64 repeat rate (95% CI) of expansions (×10−7) | (ATTCT)103 repeat rate (95% CI) of contractions (×10−7) |
| WT | 3.3 (2.4–4.1) | 6.4 (5.0–7.9) |
| Δtof1 | 17.4 (8.5–26.3) | 48.0 (35–61) |
| Δrad5 | <5 × 10−8 | 9.5 (4.0–17) |
| Δrad52 | 1.0 (0.55–1.5) | Not studied |
CI, confidence interval.
Our system also allowed us to determine the rates of repeat contractions in different genetic backgrounds. To this end, we started from a 5-FOA–resistant clone containing 103 (ATTCT)n repeats, which was obtained in the course of repeat expansion experiments, and analyzed the rate of accumulation of the URA+ clones, which could only stem from repeat contractions. All URA+ clones obtained in these experiments have shorter than 20 (ATTCT)n repeats (i.e., they resulted from large-scale contractions). Table 2 shows that the rate of contractions for the (ATTCT)103 repeat was quite low, 6.4 × 10−7 compared with other repeats studied in yeast (Table 3). In the Tof1 knockout strain, the rate of contractions was elevated 7.5-fold compared with the wild-type strain, similarly to what was observed for expansions, suggesting that the Tof1 protein acts to protect against both repeat contractions and expansions. In contrast with the expansion data, the rate of repeat contraction in the Rad5 knockout strain was unchanged. Thus, the Rad5 protein has no bearing on repeat contractions.
Table 3.
Comparison of expansion and contraction frequencies for different repeats in S. cerevisiae
| Expandable repeat | Frequency of expansions | Frequency of contractions | Reference |
| (ATTCT)64 | 1.1 × 10−6 | NS | This study |
| (GAA)100 | 1.0 × 10−5 | NS | 17 |
| (CAG)70–78 | 0.8–1.0 × 10−3 | 3.7 × 10−3 | 25, 27, 38 |
| (ATTCT)103 | NS | 2.3 × 10−6 | This study |
| (GAA)215 | NS | 1.2 × 10−2 | This study |
| (CAG)155 | 5.0 × 10−2 | 1.7 × 10−1 | 25 |
We then tested repeat-mediated fragility in tof1Δ, rad5Δ, and rad52Δ backgrounds. Unexpectedly (Fig. 5B), repeat-mediated fragility was significantly decreased (approximately threefold) in the tof1Δ strain relative to the wild-type strain for both (ATTCT)n tracts. In contrast, the absence of the Tof1 protein did not affect the fragility of the no-tract control YAC. Similar to expansions, fragility was highly dependent on the Rad5 protein, being approximately fivefold decreased in the rad5Δ background compared with wild-type levels for both repeats (Fig. 5B). Repeat-mediated fragility was slightly depressed in the rad52Δ background, but this change was not statistically different compared with wild type.
Discussion
We have shown that expansions of a pentanucleotide repeat (ATTCT)n responsible for SCA10 in humans can be observed in a yeast experimental system. The propensity of this repeat to expand depended on its length: there was an eightfold increase in the rates of expansions between the repeats differing in lengths just 1.4-fold. This observation mimics what is known about the (ATTCT)n repeat expansions in human SCA10 pedigrees (12). Admittedly, the (ATTCT)n repeat can easily expand up to thousands of copies in humans (8) but only to hundreds of copies in our yeast system. This difference is due to the fact that relatively short repeats already cause URA3 gene inactivation.
As the repeat length within the URA3 intron increased, the levels of URA3 mRNA progressively decreased. In fact, even the shortest (ATTCT)46 run studied already caused a 10-fold decrease in the URA3 mRNA level, whereas (ATTCT)81 repeat decreased it to just 2% of the control level. Furthermore, the amount of URA3 pre-mRNA downstream of the repeat also decreased dramatically with the repeat's length. In contrast, the amount of pre-mRNA upstream of the repeat was decreased insignificantly compared with the repeat-free control. Furthermore, by analyzing polyadenylated RNA transcripts via RT-PCR with an oligo(dT) primer, we showed the presence of near-identical amounts of pre-mRNA upstream of the repeat independent of its length. We conclude, therefore, that the repeat somehow signals RNA polyadenylation, resulting in the premature transcription termination and accumulation of RNA transcripts truncated at or past the repeat. The mechanisms of triggering polyadenylation by (AUUCU)n runs in yeast remain elusive. It may have to do with the fact that the polyadenylation signals are not as highly conserved in yeast as in higher eukaryotes (28). Notably, to date no decrease of pre-mRNA or processed mRNA for the mutant SCA10 allele has been detected in humans (15).
Genetic analysis of expansions revealed that knocking out the TOF1 gene leads to a fivefold increase in the rate of expansions of the (ATTCT)64 repeat compared with the wild-type strain. This effect of the Tof1 knockout on the expansions of a pentanucleotide repeat is quantitatively similar to its effect on trinucleotide repeats (17, 29). Thus, functional Tof1 protein precludes expansions of various unstable repeats. Strikingly, the deletion of the RAD5 gene in the (ATTCT)64 strain led to the complete elimination of repeat expansions. This is far more dramatic than the previous observation of a relatively modest decrease in the rate of (GAA)n repeat expansions upon RAD5 inactivation (17).
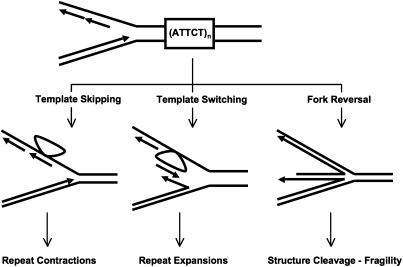
Tof1 is a fork-stabilizing protein, which in a complex with Csm3 and Mrc1 proteins prevents fork dissociation when it encounters DNA lesions or other stall sites (30). Rad5, in contrast, facilitates template switching, which allows the replication fork to bypass DNA lesions and other impediments (31). Because Rad5 seemed to be vital for the expansion of (ATTCT)n repeats, we favor a previously proposed model (17) implicating template-switch as the mechanism for expansions (Fig. 6). During the replication of repetitive tracts longer than one Okazaki fragment, the nascent leading strand might occasionally switch from its template to the nascent lagging strand, because the 3′ end of the nascent leading strand is complementary to multiple sequences in the nascent lagging strand. Upon reaching the end of an Okazaki fragment, the polymerase would have to switch back to the leading strand template for the replication to continue. After DNA replication is resumed, extra repeats remain in the nascent leading strand. When the TOF1 gene is inactivated, the replication fork becomes less stable, increasing the likelihood of template switching. Because the Rad5 protein is essential for the template switching, its inactivation should halt template switch altogether, precluding expansions.
Fig. 6.
Template skip (Left), template switch (Center), and fork reversal (Right) models for repeat contractions, expansions, and repeat-mediated fragility, respectively (see text for details).
Contractions of the (ATTCT)n repeats are also stimulated by the lack of the Tof1 protein but are independent of the Rad5 protein. This tells us that a destabilization of the replication fork, rather than template switching, is the key for repeat contractions. It is generally believed that contraction could happen when a replicative DNA polymerase skips a looped-out portion of the template strand corresponding to a repeat (Fig. 6).
(ATTCT)n repeat-mediated fragility is different from both expansions and contractions in that it is decreased upon inactivation of the TOF1 gene. This suggests that fragility occurs not during the template switching, or template skipping, but in some alternative pathway. Our data suggest that in the wild-type strain with an intact Tof1/Csm3/Mrc1 complex, template switching within the (ATTCT)n repeat is a rare event (Table 1). We suggest that fork reversal caused by either an AT-richness or a slippery nature of this repeat is more common, as was also observed for other expandable repeats (32, 33). In this scenario (Fig. 6), fragility would occur by the cleavage of reversed fork intermediates. Because Rad5 has been shown to catalyze fork reversal in vitro (34), the decrease in fragility in the rad5Δ strain would be due to a decrease in the reversed fork substrate. Alternatively, a recombination intermediate formed in the context of the PRR pathway could be a substrate for nuclease cleavage at (ATTCT) repeats, causing fragility.
The (ATTCT)n repeat differs from other expandable repeats in that it does not form stable secondary structures, such as hairpins, cruciforms, triplexes, and G-quartets (1); instead, it is a DUE (9). The proposed template-switch model does not require a repeat to form stable secondary structures. As such, it is uniquely applicable to direct tandem repeats. Note, however, that although the propensity to form alternative DNA structures is not necessary for a repeat to expand, structure-forming repeats seem to expand at a higher rate. Table 3 illustrates this point by comparing expansion and contraction rates for three repeats differing in their structure-forming ability. Evidently, the (ATTCT)n repeat has the lowest propensity to expand or contract compared with either a triplex-forming (GAA)n repeat (17) or hairpin-forming (CAG)n repeats (25, 27, 35). We believe, therefore, that the formation of stable secondary structures during replication of a repetitive sequence might additionally increase the likelihood of template switching, resulting in its higher expansion rate.
Materials and Methods
Strains.
The Saccharomyces cerevisiae strain used for the expansion assay was CH1585 (FY251) (17). The strain used for the fragility assay was BY4705 (36).
Plasmids and Selectable Cassettes.
(ATTCT)n repeats were cloned into the unique MunI site in the intron the pYES2-intron plasmid (17) (more details in SI Materials and Methods).
Gene disruption was carried out by direct gene disruption using kanMX selectable marker (details in SI Materials and Methods). Yeast genomic DNA was isolated as previously described (37).
A protocol for PCR analysis of long (ATTCT)n repeats is described in detail in SI Materials and Methods.
Rates of expansions were determined using the method of mutant accumulation (38) with the modifications described in SI Materials and Methods.
Rates of fragility were determined using fluctuation assays as previously described (25) (SI Materials and Methods).
RNA isolation and its RT-PCR analysis are described in SI Materials and Methods.
Acknowledgments
We thank Claire Moore and Mitch McVey for helpful discussions. This work was supported by the National Institutes of Health Grants GM60987 (to S.M.M.), GM063066 (to C.F.), and NS041547 (to T.A.).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1009409108/-/DCSupplemental.
References
- 1.Mirkin SM. Expandable DNA repeats and human disease. Nature. 2007;447:932–940. doi: 10.1038/nature05977. [DOI] [PubMed] [Google Scholar]
- 2.Fishel R, et al. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- 3.Jaspers NG, Gatti RA, Baan C, Linssen PC, Bootsma D. Genetic complementation analysis of ataxia telangiectasia and Nijmegen breakage syndrome: A survey of 50 patients. Cytogenet Cell Genet. 1988;49:259–263. doi: 10.1159/000132673. [DOI] [PubMed] [Google Scholar]
- 4.Niedernhofer LJ, Lalai AS, Hoeijmakers JHJ. Fanconi anemia (cross)linked to DNA repair. Cell. 2005;123:1191–1198. doi: 10.1016/j.cell.2005.12.009. [DOI] [PubMed] [Google Scholar]
- 5.Warren ST, Nelson DL. Trinucleotide repeat expansions in neurological disease. Curr Opin Neurobiol. 1993;3:752–759. doi: 10.1016/0959-4388(93)90149-s. [DOI] [PubMed] [Google Scholar]
- 6.Lenzmeier BA, Freudenreich CH. Trinucleotide repeat instability: A hairpin curve at the crossroads of replication, recombination, and repair. Cytogenet Genome Res. 2003;100:7–24. doi: 10.1159/000072836. [DOI] [PubMed] [Google Scholar]
- 7.McMurray CT. DNA secondary structure: A common and causative factor for expansion in human disease. Proc Natl Acad Sci USA. 1999;96:1823–1825. doi: 10.1073/pnas.96.5.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsuura T, et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet. 2000;26:191–194. doi: 10.1038/79911. [DOI] [PubMed] [Google Scholar]
- 9.Potaman VN, et al. Unpaired structures in SCA10 (ATTCT)n.(AGAAT)n repeats. J Mol Biol. 2003;326:1095–1111. doi: 10.1016/s0022-2836(03)00037-8. [DOI] [PubMed] [Google Scholar]
- 10.Liu G, Bissler JJ, Sinden RR, Leffak M. Unstable spinocerebellar ataxia type 10 (ATTCT*(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells. Mol Cell Biol. 2007;27:7828–7838. doi: 10.1128/MCB.01276-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato N, et al. Spinocerebellar ataxia type 31 is associated with “inserted” penta-nucleotide repeats containing (TGGAA)n. Am J Hum Genet. 2009;85:544–557. doi: 10.1016/j.ajhg.2009.09.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lin X, Ashizawa T. SCA10 and ATTCT repeat expansion: Clinical features and molecular aspects. Cytogenet Genome Res. 2003;100:184–188. doi: 10.1159/000072853. [DOI] [PubMed] [Google Scholar]
- 13.Grewal RP, et al. Clinical features and ATTCT repeat expansion in spinocerebellar ataxia type 10. Arch Neurol. 2002;59:1285–1290. doi: 10.1001/archneur.59.8.1285. [DOI] [PubMed] [Google Scholar]
- 14.Rasmussen A, et al. Clinical and genetic analysis of four Mexican families with spinocerebellar ataxia type 10. Ann Neurol. 2001;50:234–239. doi: 10.1002/ana.1081. [DOI] [PubMed] [Google Scholar]
- 15.White MC, et al. Inactivation of hnRNP K by expanded intronic AUUCU repeat induces apoptosis via translocation of PKCdelta to mitochondria in spinocerebellar ataxia 10. PLoS Genet. 2010;6:e1000984. doi: 10.1371/journal.pgen.1000984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lin X, Ashizawa T. Recent progress in spinocerebellar ataxia type-10 (SCA10) Cerebellum. 2005;4:37–42. doi: 10.1080/14734220510007897. [DOI] [PubMed] [Google Scholar]
- 17.Shishkin AA, et al. Large-scale expansions of Friedreich's ataxia GAA repeats in yeast. Mol Cell. 2009;35:82–92. doi: 10.1016/j.molcel.2009.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nedelcheva MN, et al. Uncoupling of unwinding from DNA synthesis implies regulation of MCM helicase by Tof1/Mrc1/Csm3 checkpoint complex. J Mol Biol. 2005;347:509–521. doi: 10.1016/j.jmb.2005.01.041. [DOI] [PubMed] [Google Scholar]
- 19.Tourrière H, Versini G, Cordón-Preciado V, Alabert C, Pasero P. Mrc1 and Tof1 promote replication fork progression and recovery independently of Rad53. Mol Cell. 2005;19:699–706. doi: 10.1016/j.molcel.2005.07.028. [DOI] [PubMed] [Google Scholar]
- 20.Klein HL. Reversal of fortune: Rad5 to the rescue. Mol Cell. 2007;28:181–183. doi: 10.1016/j.molcel.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 21.Hiom K. Dna repair: Rad52—the means to an end. Curr Biol. 1999;9:R446–R448. doi: 10.1016/s0960-9822(99)80278-4. [DOI] [PubMed] [Google Scholar]
- 22.Yu X, Gabriel A. Patching broken chromosomes with extranuclear cellular DNA. Mol Cell. 1999;4:873–881. doi: 10.1016/s1097-2765(00)80397-4. [DOI] [PubMed] [Google Scholar]
- 23.Radzicka A, Wolfenden R. A proficient enzyme. Science. 1995;267:90–93. doi: 10.1126/science.7809611. [DOI] [PubMed] [Google Scholar]
- 24.Freudenreich CH. Chromosome fragility: Molecular mechanisms and cellular consequences. Front Biosci. 2007;12:4911–4924. doi: 10.2741/2437. [DOI] [PubMed] [Google Scholar]
- 25.Callahan JL, Andrews KJ, Zakian VA, Freudenreich CH. Mutations in yeast replication proteins that increase CAG/CTG expansions also increase repeat fragility. Mol Cell Biol. 2003;23:7849–7860. doi: 10.1128/MCB.23.21.7849-7860.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang JH, Freudenreich CH. The Rtt109 histone acetyltransferase facilitates error-free replication to prevent CAG/CTG repeat contractions. DNA Repair (Amst) 2010;9:414–420. doi: 10.1016/j.dnarep.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sundararajan R, Gellon L, Zunder RM, Freudenreich CH. Double-strand break repair pathways protect against CAG/CTG repeat expansions, contractions and repeat-mediated chromosomal fragility in Saccharomyces cerevisiae. Genetics. 2010;184:65–77. doi: 10.1534/genetics.109.111039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zhao J, Hyman L, Moore C. Formation of mRNA 3′ ends in eukaryotes: Mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol Mol Biol Rev. 1999;63:405–445. doi: 10.1128/mmbr.63.2.405-445.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Razidlo DF, Lahue RS. Mrc1, Tof1 and Csm3 inhibit CAG.CTG repeat instability by at least two mechanisms. DNA Repair (Amst) 2008;7:633–640. doi: 10.1016/j.dnarep.2008.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Katou Y, et al. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 31.Goldfless SJ, Morag AS, Belisle KA, Sutera VA, Jr., Lovett ST. DNA repeat rearrangements mediated by DnaK-dependent replication fork repair. Mol Cell. 2006;21:595–604. doi: 10.1016/j.molcel.2006.01.025. [DOI] [PubMed] [Google Scholar]
- 32.Fouché N, Ozgür S, Roy D, Griffith JD. Replication fork regression in repetitive DNAs. Nucleic Acids Res. 2006;34:6044–6050. doi: 10.1093/nar/gkl757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kerrest A, et al. SRS2 and SGS1 prevent chromosomal breaks and stabilize triplet repeats by restraining recombination. Nat Struct Mol Biol. 2009;16:159–167. doi: 10.1038/nsmb.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Blastyák A, et al. Yeast Rad5 protein required for postreplication repair has a DNA helicase activity specific for replication fork regression. Mol Cell. 2007;28:167–175. doi: 10.1016/j.molcel.2007.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ireland MJ, Reinke SS, Livingston DM. The impact of lagging strand replication mutations on the stability of CAG repeat tracts in yeast. Genetics. 2000;155:1657–1665. doi: 10.1093/genetics/155.4.1657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brachmann CB, et al. Designer deletion strains derived from Saccharomyces cerevisiae S288C: A useful set of strains and plasmids for PCR-mediated gene disruption and other applications. Yeast. 1998;14:115–132. doi: 10.1002/(SICI)1097-0061(19980130)14:2<115::AID-YEA204>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 37.Amberg DC, Burke DJ, Strathern JN. Methods in Yeast Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab Press; 2005. [Google Scholar]
- 38.Drake JW. A constant rate of spontaneous mutation in DNA-based microbes. Proc Natl Acad Sci USA. 1991;88:7160–7164. doi: 10.1073/pnas.88.16.7160. [DOI] [PMC free article] [PubMed] [Google Scholar]