Abstract
The evolution of intimate symbiosis requires the coordination of gene expression and content between the distinct partner genomes; this coordination allows the fusion of capabilities of each organism into a single integrated metabolism. In aphids, the 10 essential amino acids are scarce in the phloem sap diet and are supplied by the obligate bacterial endosymbiont (Buchnera), which lives inside specialized cells called bacteriocytes. Although Buchnera’s genome encodes most genes for essential amino acid biosynthesis, several genes in essential amino acid pathways are missing, as are most genes for production of nonessential amino acids. Additionally, it is unresolved whether the supply of nitrogen for amino acid biosynthesis is supplemented by recycling of waste ammonia. We compared pea aphid gene expression between bacteriocytes and other body tissues using RNA sequencing and pathway analysis and exploiting the genome sequences available for both partners. We found that 26 genes underlying amino acid biosynthesis were up-regulated in bacteriocytes. Seven of these up-regulated genes fill the gaps of Buchnera’s essential amino acid pathways. In addition, genes underlying five nonessential amino acid pathways lost from Buchnera are up-regulated in bacteriocytes. Finally, our results reveal that two genes, glutamine synthetase and glutamate synthase, which potentially work together in the incorporation of ammonium nitrogen into glutamate (GOGAT) cycle to assimilate ammonia into glutamate, are up-regulated in bacteriocytes. Thus, host gene expression and symbiont capabilities are closely integrated within bacteriocytes, which function as specialized organs of amino acid production. Furthermore, the GOGAT cycle may be a key source of nitrogen fueling the integrated amino acid metabolism of the aphid–Buchnera partnership.
Keywords: Acyrthosiphon pisum, nitrogen recycling, coevolution, animal nutrition, microbiota
Animals cannot synthesize 10 amino acids and typically must obtain them from their food; these are called essential amino acids. As a diet, phloem sap is low in nitrogen and especially low in essential amino acids (1–3). Phloem-feeding insects require microbial symbionts to synthesize essential amino acids (4, 5). Conversely, these symbionts lack nonessential amino acid biosynthetic pathways; thus, the host must provide these amino acids to the symbiont (6). The best-studied model of this phenomenon is the symbiosis of the pea aphid, Acyrthosiphon pisum, and its bacterial endosymbiont, Buchnera aphidicola (4, 6–8).
Buchnera is located in the cytosol of specialized insect cells called bacteriocytes and is required for aphid survival and reproduction (4). Genomic and physiological research reveals that Buchnera synthesizes essential amino acids for its host (6, 7, 9–12). Buchnera’s genome includes genes for almost all enzymes in the essential amino acid pathways and for enzymes in pathways for the nonessential amino acids tyrosine, glycine, and cysteine (6). Despite having genes for most steps in these pathways, Buchnera is missing particular genes in five essential amino acid pathways (leucine, isoleucine, valine, methionine, and phenylalanine), raising the question of how these amino acids are produced in this symbiotic system. The A. pisum genome contains genes hypothesized to carry out the functions corresponding to these missing steps (8, 13). These genes have orthologs in other insects with sequenced genomes, in which they carry out other functions related to amino acid conversions, even though the complete pathways are absent. This widespread presence in insects and other animals makes it unclear whether their presence in the aphid genome indicates a specific role in the amino acid production capacities of bacteriocytes (8). In a study based on a limited number of expressed sequence tags (ESTs) from A. pisum bacteriocytes (14), transcripts were retrieved from only two of these complementary aphid genes (encoding enzymes 1.14.16.1 and 4.4.1.1 in the methionine and tyrosine pathways, respectively).
Another unresolved question regarding this metabolic symbiosis is the role of the host in maintaining a balanced profile of amino acids through transamination between those that are overabundant and rare. Most nitrogen available in phloem sap is in the form of free amino acids, predominantly a small subset of nonessential amino acids (15). In phloem sap directly obtained from pea aphid stylets, a few nonessential amino acids, including asparagine, glutamine, and glutamate, dominate in several host plant species used by this aphid (2). Seven nonessential amino acids (glutamate, aspartate, serine, glutamine, alanine, proline, and asparagine) are not synthesized by Buchnera; therefore, the host aphid must synthesize these amino acids and/or obtain them from the diet. Four nonessential amino acids (glutamate, aspartate, serine, and glutamine) are especially important because they are required as nitrogen-containing substrates for the synthesis of essential amino acids.
It was previously hypothesized that Buchnera recycles aphid ammonia waste into amino acids (16); however, this proposal was disproven by the publication of the Buchnera genome, which encodes neither glutamate dehydrogenase (GDH) nor glutamine synthetase (GS), the two main enzymes for incorporating ammonia into amino acids (6). Potentially, aphids themselves assimilate ammonia into amino acids, a possibility that is supported by the finding that GS, which produces glutamine from ammonia, is highly expressed in bacteriocytes (14). Glutamine has limited potential as a source of amino groups for production of other amino acids, but, together with glutamate synthase (GltS), it provides a shuttle for incorporating ammonia into most amino acids through glutamate. No gene encoding GltS was noted in the aphid genome, however (8, 13). Thus, the issue of nitrogen recycling is not resolved.
The pea aphid and its Buchnera symbiont are the animal symbiosis for which genomes of both host and symbiont are fully sequenced. We exploited these resources to address the extent to which expression of aphid genes is coordinated with the amino acid biosynthetic capabilities of Buchnera to facilitate the production of a balanced profile of amino acids from the meager and unbalanced supply present in the diet. More specifically, we used high-throughput sequencing of transcriptomes to address whether (i) aphid bacteriocyte gene expression indicates coordination with Buchnera’s essential amino acid pathways, (ii) Buchnera depends on the host to synthesize nonessential amino acids, and (iii) the bacteriocyte is able to recycle ammonia into amino acids using enzymes encoded by both genomes.
Results
Global Gene Expression in the Bacteriocyte.
Totals of 24,521,218 and 26,243,930 high-quality 74-bp reads were retrieved from mRNA obtained from aphid bacteriocytes and aphid bodies (minus bacteriocytes and embryos), respectively. Of these, 17,197,382 and 20,341,699 reads mapped to the A. pisum genome for bacteriocyte and body samples, respectively. Of 11,089 RefSeq genes, 85.7% were expressed in the bacteriocyte and 86.9% were expressed in the body. Based on cutoff criteria (twofold changes or greater and P ≤ 0.05), 404 genes were up-regulated and 996 genes were down-regulated in bacteriocytes relative to other pooled tissues (Fig. S1 and Dataset S1). The latter set included many loci involved in cuticle, nerve, and muscle production, as expected, because bacteriocytes lack these functions; down-regulated genes are not discussed further in this report. We focus on the genes up-regulated in bacteriocytes, which total only 3.6% of all RefSeq genes and 4.2% of genes expressed in our combined samples. Up-regulated genes included 26 genes encoding enzymes involved in amino acid biosynthesis; 82 genes encoding active transporters, of which 6 are annotated as amino acid transporters; and 6 immune-related genes (Dataset S2 and Table S1). Two genes (LOC100160909 and LOC100167607) showed strikingly high levels of expression in bacteriocytes, at 12,610 reads per kilobase of exon model per million mapped reads (RPKM) and 10,922 RPKM, respectively (Fig. S1 and Dataset S1). These encode conserved hypothetical proteins, one of which shows significant homology to uncharacterized proteins from other insects based on BLAST searches and one of which shows no significant homology to database proteins.
Cooperation of Aphid Genes with Buchnera’s Essential Amino Acid Pathways.
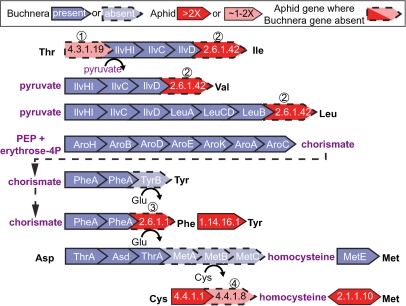
Of nine aphid genes hypothesized to contribute to the synthesis of essential amino acids, all nine are expressed in bacteriocytes and seven are significantly up-regulated greater than twofold (Fig. 1 and Table 1). The other two (LOC100165866 and LOC100159560), encoding enzymes (4.3.1.19 and 4.4.1.8, respectively) in the isoleucine and methionine pathways, are marginally up-regulated in bacteriocytes (1.75-fold and 1.4-fold, respectively). Because only ∼4% of aphid genes are up-regulated in bacteriocytes by our criteria, these results support the cooperation of aphid and symbiont gene products in the production of essential amino acids.
Fig. 1.
Complementation of host gene expression with essential amino acid pathways of Buchnera. Circled numbers indicate missing Buchnera enzymes that are complemented by up-regulated aphid enzymes. Missing Buchnera enzymes are as follows: 1, IlvA; 2, IlvE; 3, TyrB; and 4, MetC. Aphid genes with circled numbers are orthologs of missing Buchnera genes, except for 3.
Table 1.
Expression of genes underlying amino acid biosynthesis in pea aphids, comparing bacteriocytes to other body tissues
| LOC no.* | Transcript ID* | EC no.† | Product† | Pathway‡ | BAC RPKM§ | Body RPKM§ | Fold change§ | Test statistic¶ | P value¶ |
| Essential amino acid complementary genes | |||||||||
| LOC100165866 | XP_001946020.1 | 4.3.1.19 | Threonine ammonia-lyase | Ile | 244.79 | 139.76 | 1.75 | 4.80 | 0.00 |
| LOC100167587 | XP_001947354.1 | 2.6.1.42 | Branched chain-amino acid transaminase | Ile, Val, Leu | 535.92 | 113.36 | 4.73 | 15.86 | 0.00 |
| LOC100144899 | NP_001128403.1 | 2.6.1.1 | Aspartate transaminase | Phe, Tyr | 368.98 | 112.30 | 3.29 | 11.08 | 0.00 |
| LOC100161812 | XP_001943627.1 | 2.6.1.1 | Aspartate transaminase | Phe, Tyr | 0.00 | 0.00 | 0.00 | −0.21 | 1.00 |
| LOC100163139 | XP_001943882.1 | 2.6.1.1 | Aspartate transaminase | Phe, Tyr | 1.36 | 7.39 | −5.42 | −2.12 | 0.17 |
| LOC100165255 | XP_001947848.1 | 2.6.1.1 | Aspartate transaminase | Phe, Tyr | 87.41 | 114.61 | −1.31 | −2.32 | 0.11 |
| LOC100166971 | XP_001945589.1 | 1.14.16.1 | Phenylalanine 4-monooxygenase | Tyr | 869.32 | 62.33 | 13.95 | 25.58 | 0.00 |
| LOC100159197 | XP_001951155.1 | 4.4.1.1 | Similar to cystathionine β-lyase | Met | 595.31 | 79.22 | 7.51 | 19.14 | 0.00 |
| LOC100159560 | NP_001157405.1 | 4.4.1.8 | Cystathionase-like | Met | 325.10 | 236.73 | 1.37 | 3.05 | 0.02 |
| LOC100168557 | XP_001945392.1 | 2.1.1.10 | Homocysteine S-methyltransferase | Met | 145.17 | 38.78 | 3.74 | 7.46 | 0.00 |
| LOC100159972 | XP_001946395 | 2.1.1.10 | Homocysteine S-methyltransferase | Met | 13.10 | 12.18 | 1.08 | 0.04 | 1.00 |
| Nonessential amino acid aphid genes | |||||||||
| LOC100158883 | XP_001948786.1 | 1.4.1.13 | GltS | Glu | 441.30 | 75.62 | 5.84 | 15.44 | 0.00 |
| LOC100168809 | XP_001949004.1 | 2.6.1.13 | Ornithine aminotransferase | Glu | 5,603.42 | 531.70 | 10.54 | 62.69 | 0.00 |
| LOC100161005 | XP_001944430.1 | 1.5.1.2 | Pyrroline-5-carboxylate reductase | Glu | 58.00 | 73.04 | −1.26 | −1.64 | 0.39 |
| LOC100165747 | XP_001952392.1 | 1.5.1.12 | 1-Pyrroline-5-carboxylate dehydrogenase | Glu | 34.26 | 56.24 | −1.64 | −2.58 | 0.05 |
| LOC100169613 | XP_001951708.1 | 1.4.1.3 | GDH [NAD(P)+] | Glu | 67.50 | 172.43 | −2.55 | −7.22 | 0.00 |
| LOC100165735 | XP_001944345.1 | 1.1.1.95 | Phosphoglycerate dehydrogenase | Ser | 1,337.58 | 253.70 | 5.27 | 26.06 | 0.00 |
| LOC100167268 | XP_001952060.1 | 1.1.1.95 | Phosphoglycerate dehydrogenase | Ser | 86.20 | 1.67 | 51.71 | 8.75 | 0.00 |
| LOC100160394 | XP_001950951.1 | 1.1.1.95 | Phosphoglycerate dehydrogenase | Ser | 94.54 | 63.56 | 1.49 | 2.10 | 0.18 |
| LOC100168149 | XP_001948966.1 | 1.1.1.95 | Phosphoglycerate dehydrogenase | Ser | 0.00 | 0.00 | 1.00 | 0.00 | 1.00 |
| LOC100163589 | XP_001947530.1 | 2.6.1.52 | Phosphoserine transaminase | Ser | 722.28 | 118.43 | 6.10 | 20.01 | 0.00 |
| LOC100158884 | XP_001945848.1 | 3.1.3.3 | Phosphoserine phosphatase | Ser | 354.74 | 46.03 | 7.71 | 14.86 | 0.00 |
| LOC100158884 | XP_001946002.1 | 3.1.3.3 | Phosphoserine phosphatase | Ser | 354.74 | 46.03 | 7.71 | 14.86 | 0.00 |
| LOC100144899 | NP_001128403.1 | 2.6.1.1 | Aspartate transaminase | Asp | 368.98 | 112.30 | 3.29 | 11.08 | 0.00 |
| LOC100161812 | XP_001943627.1 | 2.6.1.1 | Aspartate transaminase | Asp | 0.00 | 0.00 | 0.00 | −0.21 | 1.00 |
| LOC100163139 | XP_001943882.1 | 2.6.1.1 | Aspartate transaminase | Asp | 1.36 | 7.39 | −5.42 | −2.12 | 0.17 |
| LOC100165255 | XP_001947848.1 | 2.6.1.1 | Aspartate transaminase | Asp | 87.41 | 114.61 | −1.31 | −2.32 | 0.11 |
| LOC100160265 | XP_001943429.1 | 6.3.5.4 | Asparagine synthase (glutamine-hydrolyzing) | Asp | 14.88 | 26.00 | −1.75 | −1.92 | 0.25 |
| LOC100161762 | XP_001949447.1 | 6.3.5.4 | Asparagine synthase (glutamine-hydrolyzing) | Asp | 27.09 | 25.33 | 1.07 | 0.04 | 1.00 |
| LOC100160139 | XP_001944240.1 | 6.3.1.2 | GS | Gln | 3,724.62 | 1,203.82 | 3.09 | 33.98 | 0.00 |
| LOC100165282 | XP_001947968.1 | 6.3.1.2 | Glutamate-ammonia ligase | Gln | 2,467.39 | 637.19 | 3.87 | 31.30 | 0.00 |
| LOC100163540 | XP_001947643.1 | 6.3.1.2 | Glutamate-ammonia ligase | Gln | 102.06 | 71.76 | 1.42 | 1.92 | 0.25 |
| LOC100158730 | XP_001944435.1 | 3.5.1.1 | Asparaginase | Asn | 11.16 | 18.18 | −1.63 | −1.45 | 0.51 |
| LOC100160095 | XP_001949831.1 | 3.5.1.1 | Asparaginase | Asn | 17.73 | 18.11 | −1.04 | −0.30 | 1.00 |
| LOC100167144 | XP_001942778.1 | 3.5.1.1 | Asparaginase | Asn | 26.00 | 26.51 | −1.02 | −0.28 | 1.00 |
| LOC100164899 | XP_001948711.1 | 2.6.1.2 | Alanine transaminase | Ala | 100.10 | 67.25 | 1.49 | 2.17 | 0.16 |
| LOC100163196 | XP_001947837.1 | 2.6.1.44 | Alanine-glyoxylate transaminase | Ala | 43.17 | 14.69 | 2.94 | 3.53 | 0.00 |
| LOC100165269 | XP_001949442.1 | 2.6.1.44 | Alanine-glyoxylate transaminase | Ala | 77.49 | 36.58 | 2.12 | 3.53 | 0.00 |
| LOC100164689 | XP_001944579.1 | 2.6.1.44 | Alanine-glyoxylate transaminase | Ala | 16.46 | 8.58 | 1.92 | 1.43 | 0.52 |
| LOC100161178 | XP_001948416.1 | 4.1.2.5 | Threonine aldolase | Gly | 119.47 | 52.93 | 2.26 | 4.69 | 0.00 |
| LOC100159142 | XP_001948463.1 | 4.1.2.5 | Threonine aldolase | Gly | 2.80 | 8.81 | −3.15 | −1.86 | 0.27 |
| LOC100164013 | XP_001943196.1 | 1.4.4.2 | Glycine dehydrogenase (decarboxylating) | Gly | 646.66 | 54.62 | 11.84 | 21.61 | 0.00 |
| LOC100168482 | XP_001943549.1 | 2.1.2.10 | Aminomethyltransferase | Gly | 459.77 | 34.33 | 13.39 | 18.51 | 0.00 |
| LOC100162429 | XP_001948247.1 | 1.8.1.4 | Dihydrolipoyl dehydrogenase | Gly | 358.57 | 270.51 | 1.33 | 2.79 | 0.03 |
Bold EC numbers represent up-regulated bacteriocyte genes relative to the body, wherein P < 0.05 and the bacteriocyte fold change is ≥2.
*NCBI-designated LOC numbers and transcript IDs for Acyr_1.0.
†Enzyme Commission (EC)-designated enzyme numbers and product names from the BRENDA Enzyme Information System and NCBI.
‡Amino acid pathway enzyme participates in from Kyoto Encyclopedia of Genes and Genomes and the MetaCyc Encyclopedia of Metabolic Pathways.
§Bacteriocyte (BAC) and body gene expression in RPKM.
¶Kal et al.’s Z-test statistics and false discovery rate-corrected P values for comparing BAC and body LOC RPKM.
Buchnera Requires the Host to Synthesize Nonessential Amino Acids.
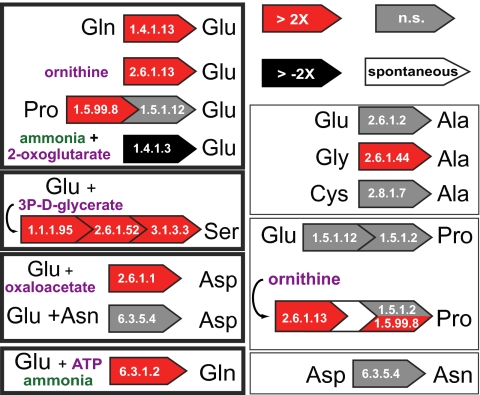
Genes underlying biosynthesis of five of the seven nonessential amino acids not synthesized by Buchnera (glutamate, aspartate, serine, glutamine, and alanine) are up-regulated in the bacteriocyte relative to the body (Fig. 2 and Table 1). Two pathways for nonessential amino acids (proline and asparagine) are not up-regulated in the bacteriocyte (Fig. 2 and Table 1).
Fig. 2.
Relative expression in bacteriocytes of A. pisum genes underlying biosynthesis of nonessential amino acids not synthesized by Buchnera. Enzyme Commission numbers are given to indicate enzymatic steps in pathways. Boxes outlined in bold indicate nonessential amino acids required by Buchnera to synthesize essential amino acids for the host. Spontaneous indicates a spontaneous reaction not requiring an enzyme. n.s., not significant.
Recycling of Waste Ammonia into Amino Acids in Aphid Bacteriocytes.
Genes for two aphid enzymes that assimilate ammonia into amino acids are significantly up-regulated greater than twofold in bacteriocytes. One aphid enzyme (4.4.1.1 encoded by LOC100159197) is a putative cystathionine γ-lyase, which assimilates 2-oxobutanoate and ammonia into l-cystathionine as part of the methionine pathway (Fig. 1 and Table 1). This may contribute to methionine or cysteine production but is not expected to form a significant contribution to the amino acid budget, because cystathionine or methionine is not a substrate for making other amino acids. GS (6.3.1.2) uses ammonia and glutamate to produce glutamine, and both aphid gene copies (LOC100160139 and LOC100165282) encoding GS are significantly up-regulated (at 3.1-fold and 3.9-fold, respectively) in aphid bacteriocytes (Fig. 2 and Table 1). Up-regulation of GS in bacteriocytes verifies previous findings based on limited numbers of ESTs and on quantitative PCR (14). Glutamine can contribute amino groups during synthesis of arginine and histidine. GS alone is not expected to enable significant recycling of amino acids, however, because glutamine is not a substrate for most amino acid biosynthetic pathways.
Another gene found to be up-regulated was LOC100158883, at 5.8-fold higher expression in bacteriocytes (Fig. 2 and Table 1). BLAST searches reveal that this gene encodes GltS, which produces two glutamates from glutamine. The finding that both GS and GltS are highly up-regulated in bacteriocytes suggests that aphid-encoded enzymes can recycle waste ammonia for the production of glutamate (Fig. 3). Another major substrate for the essential amino acid pathways encoded by Buchnera is aspartate, and the bacteriocytes show strong up-regulation of LOC100144899 (2.6.1.1), encoding glutamyl-aspartate transferase and enabling interconversion of aspartate and glutamate (Fig. 2 and Table 1). In addition, LOC100168809 (2.6.1.13) is up-regulated in the bacteriocyte and is an ornithine-oxoacid transaminase that interconverts ornithine and glutamate, wherein ornithine is an important intermediate in the arginine biosynthetic pathway encoded by Buchnera (Fig. 4 and Table 1). Overall, bacteriocytes show gene expression patterns expected to promote the recycling of waste nitrogen into substrates needed for essential amino acid production by Buchnera.
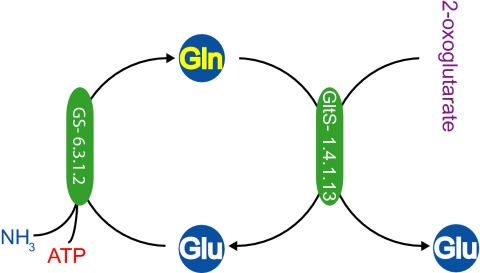
Fig. 3.
Up-regulation of genes underlying the GOGAT cycle in aphid bacteriocytes relative to other tissues.
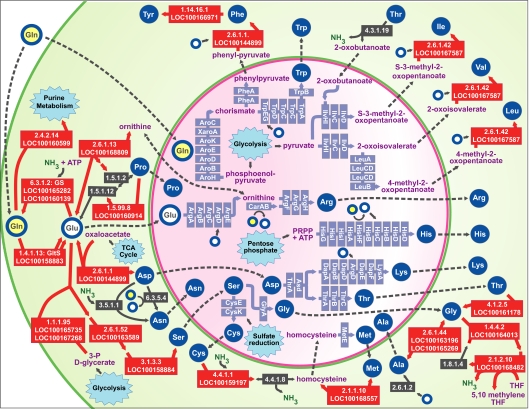
Fig. 4.
Integration of amino acid biosynthetic pathways of the aphid and Buchnera within the aphid bacteriocyte. Green and pink areas represent cytosol of the bacteriocyte and of a Buchnera cell, respectively. Green and pink lines represent aphid and Buchnera membranes, respectively. Amino acids are represented by blue disks or by yellow (glutamine) or white (glutamate) disks with blue outlines. Aphid genes are represented by red (up-regulated in bacteriocyte) or brown (not significantly up-regulated) boxes labeled with Enzyme Commission numbers and LOC numbers corresponding to NCBI annotation of the A. pisum genome. Purple boxes represent Buchnera genes.
Discussion
The amino acid metabolism in this symbiotic system consists of an integrated network involving gene products of both the aphid host and its symbiont. Our results support several ways in which processes encoded by both partner genomes are integrated: complementarity of genomes in encoding enzymes within several pathways, complementarity of nonessential and essential amino acid biosynthetic pathways encoded by both genomes, amino transferase activities that allow reallocation of amino nitrogen among end products, and recycling of waste ammonia to enhance the amino acid supply.
First, our results suggest that aphid-encoded gene products fill gaps in enzymatic pathways encoded by the Buchnera genome. Genes are missing from each of six essential amino acid pathways (for isoleucine, valine, leucine, phenylalanine, tyrosine, and methionine); the corresponding genes are up-regulated in the bacteriocyte relative to other tissues and cells (Figs. 1 and 4 and Table 1). B. aphidicola strains from divergent aphid species lack the same genes in these pathways (17, 18), suggesting their loss early in the evolution of this symbiosis.
Second, bacteriocyte gene expression reveals complementarity between amino acid pathways encoded by the two genomes. All seven nonessential amino acid pathways missing from Buchnera are up-regulated in the bacteriocyte relative to the body, except for pathways for biosynthesis of proline and asparagine (Figs. 2 and 4 and Table 1). Based on RPKM values, both genes (1.5.1.12 and 1.5.1.2) in the proline pathway are highly expressed in the body and bacteriocyte (Table 1), suggesting that proline is being synthesized at substantial levels. Of the two copies of the gene corresponding to the single step in the asparagine pathway (6.3.5.4), both copies are weakly expressed (Table 1). Because asparagine is the dominant amino acid found in phloem of A. pisum host plants (2, 19, 20), it is expected to be present in excess, making its biosynthesis unimportant. All four nonessential amino acid pathways (glutamate, glutamine, aspartate, and serine) that generate required amino group donors for Buchnera’s essential amino acid pathways are significantly up-regulated in the bacteriocyte relative to the body (Fig. 2 and Table 1).
A key question is which amino acids are imported into the bacteriocyte from the body to fuel up-regulated amino acid biosynthesis pathways. Previous studies (21, 22) showed that glutamine and asparagine are the most abundant amino acids in pea aphid hemolymph. Because phloem concentrations of amino acids do impact hemolymph amino acid levels (23), this potentially results from the high concentrations of these two amino acids in ingested phloem (2, 19, 20) but may also reflect their generation within aphid tissues. Ammonia waste from aphid tissues may be used for glutamine production via GS, with the resulting glutamine imported into bacteriocytes, where it can contribute to glutamate production. Based on radiochemical labeling, glutamine but not glutamate is actively taken into bacteriocytes, whereas glutamate but not glutamine is actively taken into Buchnera cells (21). In light of Buchnera’s biosynthetic capacities (6), radiochemical labeling (21), and our transcriptome data, glutamine may be the primary amino acid imported into bacteriocytes and glutamate is the primary amino acid imported into Buchnera cells. Based on our transcriptome data and on Buchnera’s gene set, additional amino acids, such as glutamine, aspartate, and serine, must also be imported into Buchnera cells to serve as substrates for essential amino acid biosynthesis (Fig. 4).
Third, our results suggest that aphid-encoded enzymes within the bacteriocyte serve to adjust the profile of nonessential amino acids to fit the needs of both Buchnera and the host. Nitrogen in the form of glutamate and glutamine is readily available for metabolic processes because these amino acids are important substrates for the synthesis of amino acids, nucleic acids, and other nitrogen-containing compounds (24). We found that three genes encoding glutamate aminotransferases (2.6.1.52, 2.6.1.1, and 2.6.1.42) are up-regulated in aphid bacteriocytes (Table 1). These aminotransferases provide the source of amino groups for seven amino acid pathways for which underlying genes are also up-regulated in the bacteriocyte (serine, aspartate, phenylalanine, tyrosine, isoleucine, valine, and leucine) (Fig. 4). Glutamate is also required as a substrate for Buchnera’s glutamate aminotransferases in the arginine, tryptophan, and lysine pathways. Moreover, glutamate is an indirect amino group donor in the synthesis of glycine, cysteine, lysine, and threonine via up-regulated bacteriocyte genes in the pathways for Ser and Asp (Fig. 4). Glutamine is the amino group donor in the bacteriocyte for the up-regulated aphid enzyme (2.4.2.14) underlying purine synthesis (Fig. 4). Glutamine is also a direct amino group donor in Buchnera’s arginine and histidine pathways (Fig. 4). In summary, our results suggest that the bacteriocyte requires glutamine and especially glutamate for the synthesis of amino acids and for purine metabolism. In turn, regulation of glutamate and glutamine levels in the bacteriocyte is important for maintaining a balanced profile of amino acids for the aphid host and Buchnera.
Finally, our results support a possible role of the bacteriocyte in recycling ammonia waste for the production of glutamine and glutamate, which, in turn, contribute to the synthesis of most other amino acids via pathways encoded in either the aphid or Buchnera genome. The overall level of amino nitrogen is limiting to aphid growth, because phloem sap has a very high carbon/nitrogen ratio. Past studies have yielded mixed results regarding whether nitrogen recycling occurs in aphids or Buchnera (16, 25). Based on a nitrogen isotope study, N-15 glutamine is converted into N-15 glutamate in the bacteriocyte cytosol (21). Sasaki and Ishikawa (21) hypothesized that enzyme 3.5.1.2. (glutaminase) is primarily responsible for converting glutamine into glutamate and ammonia in the bacteriocyte and that ammonia is converted subsequently into more glutamate via glutamine dehydrogenase (GDH). The aphid genome and our results weigh against this hypothesis, because the aphid genome does not encode glutaminase and GDH is down-regulated in bacteriocytes. A second hypothesis that Sasaki and Ishikawa (21) proposed is that glutamine is converted into glutamate in the bacteriocyte via enzyme 1.4.1.13 (GltS); however, they dismissed this hypothesis based on evidence at that time that this enzyme is not widely found in animals. However, GltS is present in the pea aphid genome and is significantly up-regulated in the bacteriocyte.
We propose that both ammonia recycling and the maintenance of glutamine and glutamate in the bacteriocyte rely on both GS and GltS, which are up-regulated in the bacteriocyte (Table 1). GS was found to be highly expressed in bacteriocytes in our study as well as in a previous study (14), but this finding could reflect a role in detoxification of ammonia waste rather than a substantial role in supplementing the amino acid budget. Our results also show elevated expression of GltS, which was not previously detected in bacteriocyte ESTs. The elevated expression of both GS and GltS in the bacteriocyte suggests substantial levels of ammonia assimilation. In plants and most microbes, these two enzymes act together to form a shuttle for incorporation of ammonium nitrogen into glutamate (GOGAT cycle) (26) (Fig. 3), the substrate for the majority of amino acid pathways. GDH, an alternative to GS/GltS as an enzyme for ammonia assimilation into glutamate, is present in aphids but is down-regulated in bacteriocytes (Table 1). GDH in body tissues is likely involved in generation of ammonia from glutamate, the activity considered as being usual in animal tissues. GS/GltS is the dominant mode of ammonia assimilation when glucose is plentiful (because GS is ATP-dependent) and ammonia is in low concentration (27–29). These conditions apply to aphids, which ingest excess sugar and must maintain ammonia at low concentrations in cells because of its toxicity. Furthermore, the bacteriocyte itself is likely high in glucose, because the enzyme (trehalase) converting trehalose (the major carbohydrate circulating in insect hemolymph) to glucose (30) is significantly up-regulated in the bacteriocyte relative to the body (LOC100162689, Dataset S1). Ample glucose in bacteriocytes can fuel glycolysis in both Buchnera and the bacteriocyte, resulting in abundant ATP and favoring the GS/GltS cycle. The quantitative contribution of this cycle to amino nitrogen supply is not resolved, however.
Among other insects, the GS/GltS cycle has been characterized in vivo in mosquitoes (31, 32) and silkworms (33–35). In the silkworm, GltS activity is associated with nitrogen recycling through the GS/GltS cycle; GltS activation in the posterior silk glands allows enhanced utilization of nitrogen (in the form of glutamate converted from glutamine) for the synthesis of silk protein (33–35). In the mosquito, GS/GltS activity is linked to ammonia detoxification; following a blood meal, GS and GltS are highly expressed in the fat body (32), the main tissue involved in ammonia detoxification (36).
Also important in facilitating the interaction with Buchnera are genes related to transport and immunity. Our expression data suggest that transport of amino acid-related metabolites and substrates across both host-derived membranes in the bacteriocyte (the bacteriocyte cell membrane and the membrane encasing individual Buchnera) (Fig. 4) is required. A total of 82 active transporters were up-regulated in the bacteriocyte, and these include amino acid, sugar, organic acid, and ABCC and ABCG type transporters (Dataset S2). Four of six amino acid transporters up-regulated were cationic amino acid transporters (Dataset S2). This type of transporter is responsible for transporting positively charged amino acids, such as Buchnera-synthesized histidine, lysine, and arginine, from one side of the membrane to the other. Two other types of amino acid transmembrane transporters were also upregulated in the bacteriocyte (Dataset S2) and may play an important role in exporting amino acids outside of the bacteriocyte to the hemolymph as well as transporting amino acids across the Buchnera-enclosed host membrane. Six immune-related genes were up-regulated in the bacteriocyte (Dataset S2). These include one GST and two lysozyme-related enzymes (Dataset S2), which are known to detoxify stress-causing agents and to degrade bacterial cell walls, respectively (37). Potentially, lysozyme and detoxification enzymes help to defend against invading microbes. Two immune genes belonging to the JAK/STAT pathway and one gene belonging to the JNK pathway were also up-regulated (Dataset S2). The roles of these pathways are presently unknown.
In conclusion, global gene expression of the bacteriocyte reveals that the pea aphid and Buchnera cooperate to synthesize a full spectrum of required amino acids. Although evidence based on more limited sequencing of bacteriocytes shows some elevated expression of amino acid-related genes in pea aphid bacteriocytes (14), this study provides support for a genome-wide coordination of host gene expression with bacterial metabolic pathways in a host–endosymbiont symbiosis. Buchnera produces essential amino acids that are deficient in the aphid's diet with the help of complementary aphid-encoded enzymes. Nonessential amino acids that are not synthesized by Buchnera are synthesized by the aphid's bacteriocyte. Maintenance of a balanced profile of amino acids by the aphid is most likely achieved by the generation of glutamate for the anabolism of other amino acids through the GOGAT cycle. Ammonia recycling via the GOGAT cycle in the bacteriocyte may be a key mechanism in sustaining this nitrogen-limited symbiotic relationship.
Materials and Methods
Aphid Rearing, Dissections, and Extraction.
A. pisum strain LSR1 was used in this study. Aphids were reared in “cup cages” on broad bean, Vicia faba (38). More details on aphid rearing and dissections are provided in SI Materials and Methods.
RNAseq Analysis and Statistics.
Illumina library preparation and sequencing were conducted by Yale University's Keck Genome Sequencing Center. Libraries were sequenced as 74-mers using Illumina’s pipeline. One sample was sequenced per lane. Reads for both RNAseq samples were submitted to the Gene Expression Omnibus (GEO) database of the National Center for Biotechnology Information (NCBI) (accession no. SRA023619.1). RNASeq data were mapped to A. pisum’s genome, version Acyr_1.0, with 11,089 RefSeq genes and were annotated using the CLC bio Genomics Workbench. Normalized gene locus expression levels were calculated as RPKM (39). Statistical comparison of RPKM values between samples was conducted by Kal et al.’s test (Z test) (40), and multiple comparisons were conducted with a false discovery rate criterion using the Genomics Workbench. Statistical significance was determined if P ≤ 0.05 and when a greater than twofold change in expression of the bacteriocyte relative to the body occurred. Aphid amino acid pathways and putative enzyme functions were analyzed using the A. pisum genome and annotations in the NCBI, Kyoto Encyclopedia of Genes and Genomes (KEGG) (41), BRENDA Enzyme Information System (42), EcoCyc (43), and AcypiCyc (44) databases.
Acknowledgments
We are grateful to Kim Hammond for greenhouse help, Kevin Vogel for bacteriocyte dissection help, and Becky Nankivell for help with figures. We also thank two anonymous reviewers for their valuable comments and critiques, which improved this manuscript. This work was supported by National Science Foundation Award 0723472 (to N.A.M.).
Footnotes
The authors declare no conflict of interest.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. SRA023619.1).
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013465108/-/DCSupplemental.
References
- 1.Slansky FJ, Scriber JM. Food consumption and utilization. In: Kerkut GA, Gilbert LI, editors. Comprehensive Insect Physiology, Biochemistry and Pharmacology. Vol. 4. Oxford: Pergamon; 1985. pp. 87–163. [Google Scholar]
- 2.Sandström J, Pettersson J. Amino acid composition of phloem sap and the relation to intraspecific variation in pea apid (Acyrthosiphon pisum) performance. J Insect Physiol. 1994;40:947–955. [Google Scholar]
- 3.Sandström J, Moran N. Amino acid budgets in three aphid species using the same host plant. Physiol Entomol. 1999;26:202–211. [Google Scholar]
- 4.Baumann P. Biology bacteriocyte-associated endosymbionts of plant sap-sucking insects. Annu Rev Microbiol. 2005;59:155–189. doi: 10.1146/annurev.micro.59.030804.121041. [DOI] [PubMed] [Google Scholar]
- 5.Buchner P. Endosymbiosis of Animals with Plant Microorganisms. New York: Interscience; 1965. p. 909. [Google Scholar]
- 6.Shigenobu S, Watanabe H, Hattori M, Sakaki Y, Ishikawa H. Genome sequence of the endocellular bacterial symbiont of aphids Buchnera sp. APS. Nature. 2000;407:81–86. doi: 10.1038/35024074. [DOI] [PubMed] [Google Scholar]
- 7.Douglas AE, Prosser WA. Synthesis of the essential amino acid tryptophan in the pea aphid (Acyrthosiphon pisum) symbiosis. J Insect Physiol. 1992;38:565–568. [Google Scholar]
- 8.Wilson ACC, et al. Genomic insight into the amino acid relations of the pea aphid, Acyrthosiphon pisum, with its symbiotic bacterium Buchnera aphidicola. Insect Mol Biol. 2010;19(Suppl 2):249–258. doi: 10.1111/j.1365-2583.2009.00942.x. [DOI] [PubMed] [Google Scholar]
- 9.Douglas AE. Sulphate utilisation in an aphid symbiosis. Insect Biochem. 1988;18:599–605. [Google Scholar]
- 10.Sasaki T, Fukuchi N, Ishikawa H. Amino acid flow through aphid and its symbiont: Study with 15N-labeled glutamine. Zoolog Sci. 1993;10:787–791. [Google Scholar]
- 11.Baumann P, et al. Genetics, physiology, and evolutionary relationships of the genus Buchnera: Intracellular symbionts of aphids. Annu Rev Microbiol. 1995;49:55–94. doi: 10.1146/annurev.mi.49.100195.000415. [DOI] [PubMed] [Google Scholar]
- 12.Febvay G, Liadouze I, Guillaud J, Bonnot G. Analysis of energetic amino acid metabolism in Acyrthosiphon pisum: A multidimensional approach to amino acid metabolism in aphids. Arch Insect Biochem Physiol. 1995;29:45–69. [Google Scholar]
- 13.International Aphid Genomics Consortium Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 2010;8:e1000313. doi: 10.1371/journal.pbio.1000313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakabachi A, et al. Transcriptome analysis of the aphid bacteriocyte, the symbiotic host cell that harbors an endocellular mutualistic bacterium, Buchnera. Proc Natl Acad Sci USA. 2005;102:5477–5482. doi: 10.1073/pnas.0409034102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sandström J, Moran N. How nutritionally imbalanced is phloem sap for aphids? Entomol Exp Appl. 1999;91:203–210. [Google Scholar]
- 16.Whitehead LF, Wilkinson TL, Douglas AE. Nitrogen recycling in the pea aphid symbiosis. Proc Biol Sci. 1992;250:115–117. [Google Scholar]
- 17.Moran NA, McCutcheon JP, Nakabachi A. Genomics and evolution of heritable bacterial symbionts. Annu Rev Genet. 2008;42:165–190. doi: 10.1146/annurev.genet.41.110306.130119. [DOI] [PubMed] [Google Scholar]
- 18.Moran NA, McLaughlin HJ, Sorek R. The dynamics and time scale of ongoing genomic erosion in symbiotic bacteria. Science. 2009;323:379–382. doi: 10.1126/science.1167140. [DOI] [PubMed] [Google Scholar]
- 19.Sasaki T, Aoki T, Hayashi H, Ishikawa H. Amino acid composition of the honeydew of symbiotic and aposymbiotic pea aphids Acyrthosiphon pisum. J Insect Physiol. 1990;36:35–40. [Google Scholar]
- 20.Akman Gündüz E, Douglas AE. Symbiotic bacteria enable insect to use a nutritionally inadequate diet. Proc Biol Sci. 2009;276:987–991. doi: 10.1098/rspb.2008.1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sasaki T, Ishikawa H. Production of essential amino acids from glutamate by mycetocyte symbionts of the pea aphid, Acyrthosiphon pisum. J Insect Physiol. 1995;41:41–46. [Google Scholar]
- 22.Sasaki T, Ishikawa H. Nitrogen recycling in the endosymbiotic system of the pea aphid, Acyrthosiphon pisum. Zoolog Sci. 1993;10:779–785. [Google Scholar]
- 23.Moran NA, Dunbar HE, Wilcox JL. Regulation of transcription in a reduced bacterial genome: Nutrient-provisioning genes of the obligate symbiont Buchnera aphidicola. J Bacteriol. 2005;187:4229–4237. doi: 10.1128/JB.187.12.4229-4237.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klowden M. Physiological Systems in Insects. San Diego: Academic; 2002. [Google Scholar]
- 25.Wilkinson TL, Douglas AE. Why aphids lacking symbiotic bacteria have elevated levels of the amino acid glutamine. J Insect Physiol. 1995;41:921–927. [Google Scholar]
- 26.Buchanan B, Gruissem W, Jones R. Biochemistry and Molecular Biology of Plants, American Society of Plant Physiologists. Waldorf, MD: Courier Companies, Inc.; 2000. [Google Scholar]
- 27.Helling R. Pathway choice in glutamate synthesis in Escherichia coli. J Bacteriol. 1998;17:4571–4575. doi: 10.1128/jb.180.17.4571-4575.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Drews M, et al. Pathways of glutamine metabolism in Spodoptera frugiperda (Sf9) insect cells: Evidence for the presence of the nitrogen assimilation system, and a metabolic switch by 1H/15N NMR. J Biotechnol. 2000;78:23–37. doi: 10.1016/s0168-1656(99)00231-x. [DOI] [PubMed] [Google Scholar]
- 29.Campbell MK, Farrell SO. Biochemistry. 5th Ed. Belmont, CA: Thompson Brooks/Cole; 2006. [Google Scholar]
- 30.Becker A, Schlöder P, Steele JE, Wegener G. The regulation of trehalose metabolism in insects. Experientia. 1996;52:433–439. doi: 10.1007/BF01919312. [DOI] [PubMed] [Google Scholar]
- 31.Scaraffia PY, Zhang Q, Wysocki VH, Isoe J, Wells MA. Analysis of whole body ammonia metabolism in Aedes aegypti using [15N]-labeled compounds and mass spectrometry. Insect Biochem Mol Biol. 2006;36:614–622. doi: 10.1016/j.ibmb.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 32.Scaraffia PY, Zhang Q, Thorson K, Wysocki VH, Miesfeld RL. Differential ammonia metabolism in Aedes aegypti fat body and midgut tissues. J Insect Physiol. 2010;56:1040–1049. doi: 10.1016/j.jinsphys.2010.02.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shinbo H, Konno K, Hirayama C. The pathway of ammonia assimilation in the silkworm, Bombyx mori. J Insect Physiol. 1997;43:959–964. doi: 10.1016/s0022-1910(97)00045-0. [DOI] [PubMed] [Google Scholar]
- 34.Hirayama C, Saito H, Konno K, Shinbo H. Purification and characterization of NADH-dependent glutamate synthase from the silkworm fat body (Bombyx mori) Insect Biochem Mol Biol. 1998;28:473–482. doi: 10.1016/s0965-1748(98)00019-8. [DOI] [PubMed] [Google Scholar]
- 35.Hirayama C, Nakamura M. Regulation of glutamine metabolism during the development of Bombyx mori larvae. Biochim Biophys Acta. 2002;1571:131–137. doi: 10.1016/s0304-4165(02)00207-6. [DOI] [PubMed] [Google Scholar]
- 36.Scaraffia PY, Isoe J, Murillo A, Wells MA. Ammonia metabolism in Aedes aegypti. Insect Biochem Mol Biol. 2005;35:491–503. doi: 10.1016/j.ibmb.2005.01.012. [DOI] [PubMed] [Google Scholar]
- 37.Gerardo NM, et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11:R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Oliver KM, Russell JA, Moran NA, Hunter MS. Facultative bacterial symbionts in aphids confer resistance to parasitic wasps. Proc Natl Acad Sci USA. 2003;100:1803–1807. doi: 10.1073/pnas.0335320100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mortazavi A, Williams BA, McCue K, Schaeffer L, Wold B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat Methods. 2008;5:621–628. doi: 10.1038/nmeth.1226. [DOI] [PubMed] [Google Scholar]
- 40.Kal AJ, et al. Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell. 1999;10:1859–1872. doi: 10.1091/mbc.10.6.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: New content and tools in 2009. Nucleic Acids Res. 2009;37(Database issue):D588–D592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Keseler IM, et al. EcoCyc: A comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37(Database issue):D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.AcypiCyc AcypiCyc database, NRIA Bamboo team-project. Database is hosted at the Pôle Rhône-Alpes de Bioinformatique (PRABI) 2010. Available at http://acypicyc.cycadsys.org/ Accessed June 1, 2010.