Abstract
Recent studies have suggested that Drosophila taste receptors are encoded by a family of G protein-coupled receptor genes comprising at least 56 members. One of these genes, Gr5a, has been shown by genetic analysis to be required by the fly for behavioral and sensory responses to a sugar, trehalose. Here, we show that Gr5a is expressed in neurons of taste sensilla located on the labellum and legs. Expression is observed in most if not all labellar sensilla and suggests that many taste neurons express more than one receptor. We demonstrate by heterologous expression in a Drosophila S2 cell line that Gr5a encodes a receptor tuned to trehalose. This is the first functional expression of an invertebrate taste receptor.
The majority of taste sensilla in Drosophila are located on the labellum, a gustatory organ of the proboscis, and the tarsal segments of the legs. Most sensilla house four gustatory neurons, one sensitive to sugars, one strongly sensitive to salt, one weakly sensitive to salt, and one responsive to water, as well as one mechanosensory neuron (1, 2). Previous work has identified a large family of G protein-coupled receptor genes, the Gr genes, which have been proposed to encode gustatory receptors in Drosophila (3). The Gr family comprises at least 56 members, many of which are expressed in subsets of taste neurons in the different taste organs (3-5).
Perception of sugars is a critical taste modality in animals such as fruit flies that ingest sweet substances for nutrition. Among the sugars, one that plays a particularly important role in insects is the disaccharide trehalose [1-O-(α-d-glucopyranosyl-α-d-glucopyranose)], which is composed of two glucose molecules connected by an unusual 1,1′ linkage. Also called mycose, this disaccharide is abundant in yeasts and fungi, which are present in fermented fruit, an important food source of Drosophila. Trehalose is especially critical to insects in that it is the principal sugar found in the hemolymph, where it is involved in regulation of osmolarity, and in many winged insects it is an easily transported and accessible energy source metabolized during flight (6).
Previous genetic studies have identified a locus on the X chromosome called Tre, whose alleles confer differing levels of sensitivity to trehalose (7, 8). Recently we have shown that a member of the Gr family, Gr5a, maps to this locus and is necessary for trehalose response in vivo (9). Deletion mutants of Gr5a have a greatly diminished response to trehalose when assayed by electrophysiological recordings from single taste sensilla, or by a behavioral test. This defect was rescued by reintroducing a functional copy of Gr5a on a transgene, but not by introducing a mutant copy of Gr5a. Consistent with our results, Ueno et al. (10) showed that polymorphisms in Gr5a correlate with trehalose responses. However, these results do not exclude the possibility that Gr5a plays an indirect role in trehalose response.
Here we show that Gr5a is expressed in taste neurons of the labellum and the tarsi, supporting its identity as a taste receptor gene. We then provide direct evidence that Gr5a encodes a trehalose receptor by expressing it in Drosophila S2 cells and showing that stimulation with trehalose evokes changes in intracellular calcium (Ca2+) levels in these cells. We further show that Gr5a is narrowly tuned to trehalose, showing little if any response to other disaccharides.
Methods
Expression Analysis. An 8.5-kb fragment upstream of Gr5a was amplified from genomic DNA of Canton-S flies and inserted upstream of the GAL4-coding sequence in the pG4PN vector (C. Warr and J.R.C., unpublished data). Transgenic flies were generated by using standard procedures. Heterozygous flies carrying one copy each of Gr5a-GAL4 and UAS-lacZ were stained for LacZ activity. For visualization of GFP, flies carrying two copies of Gr5a-GAL4, as well as two copies of UAS-mCD8:GFP, were examined by using confocal microscopy.
Heterologous Expression. A 1.2-kb fragment of full-length Gr5a cDNA was amplified from head mRNA by using the Smart RACE cDNA Amplification kit (Clontech). This fragment was inserted into a unique EcoRI site in the pRmHa3 vector (11). Drosophila S2 cells were grown at 25°C in Shields and Sang M3 insect medium supplemented with 10% FCS and antibiotic/antimycotic solution. S2 cells were cotransfected with pRmHa3-Gr5a and pIZT-V5/His vector (Invitrogen) encoding GFP and zeocin-resistance to facilitate recognition of transfectants and antibiotic selection, at a 3:1 ratio by using liposomal formulation (CellFectin, Invitrogen) according to the manufacturer's instructions. Expression of Gr5a was induced by adding 0.6 mM Cu2+ to the cell culture media 48 hrs before an experiment. Levels of Gr5a expression in uninduced and induced cells were examined by RT-PCR to confirm the induction of Gr5a.
Calcium Imaging. Transfected cells were grown in 96-well plates and loaded with membrane-permeable fura 2-acetoxymethyl ester (fura 2-AM) as described elsewhere (12). Briefly, after being washed in Hanks' balanced salt solution (HBSS), cells were incubated for 1 h with 1-2 μM fura 2-AM (in 10% Pluronic F-127) at room temperature and under low light conditions. Subsequently, fura 2-AM solution was removed and cells were incubated for 1 h in HBSS (50 μl) to allow endogenous esterases to cleave AM ester. Cells were stimulated by addition of 50 μl of 2× tastant solution. Response kinetics were measured from cells loaded with 100 μM fura 2 via patch pipette and stimulated with 1× tastant solution via a puffer pipette under control of PicoPump 830 (World Precision Instruments, Sarasota, FL). All chemicals were purchased from Sigma. Imaging experiments were conducted on a Nikon TE300 inverted microscope equipped with a Nikon superfluor lens (×10/0.5 and ×20/0.75). Cells were stimulated with 340 and 380 nm UV light from a DeltaRAM monochromator, and resulting images were collected by using an IC-200 intensified charge-coupled device camera; data acquisition and analysis were performed by using an ImageMaster suite (PTI, South Brunswick, NJ). Individual responses were recorded for 60 s. F340/380 ratios were analyzed to measure Ca2+ release. A responder cell was defined as one showing a ratio of 0.34 or above. In general, >90% of responses showed a ratio of at least 0.51.
Results
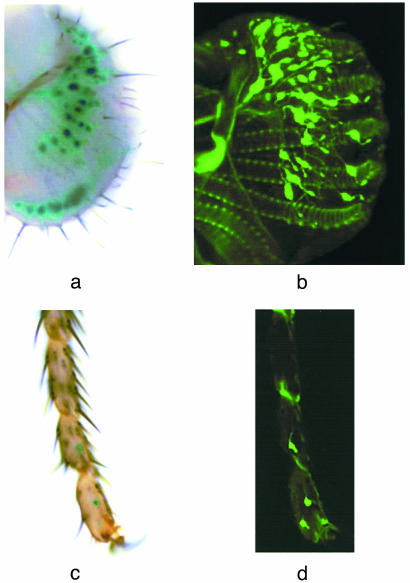
To test the hypothesis that Gr5a plays a direct role in trehalose reception, we first asked whether it is expressed in taste neurons. Because previous attempts at in situ hybridization have proved unsuccessful with the great majority of Gr genes (3, 4) we generated Gr5a promoter-GAL4 lines. An 8.5-kb genomic region upstream of Gr5a was used to supply a promoter, and GAL4 was used to drive expression of both UAS-lacZ and UAS-GFP reporters. We observed wide expression in taste neurons of the labellum as well as in four to six neurons in the tarsi of adult flies (Fig. 1). No sexual dimorphism was observed in the expression pattern. Six independently derived lines were examined and all gave equivalent results.
Fig. 1.
Expression of Gr5a in taste neurons in the labellum and distal segments in the leg. Shown here are whole mount samples of labella or tarsi. Genotypes examined were as follows: Gr5a-GAL4/+; UAS-lacZ/+ (a and c) and Gr5a-GAL4/UAS-mCD8:GFP; Gr5a-GAL4/UAS-mCD8:GFP (b and d). Shown in b is a composite of a series of confocal images. Reporter gene expression was observed in ≈30 cells in each half of the labellum and 2 cells in each of the two to three distal-most segments of the leg.
To examine Gr5a function at the cellular level, we expressed Gr5a cDNA in Drosophila S2 cells. This cell line was chosen for two reasons. First, chemosensory receptors have been notoriously difficult to express in heterologous systems and we predicted that use of a Drosophila cell line might improve the possibility of functional expression of a Drosophila receptor. Second, previous studies have documented Ca2+ release after activation of G protein-coupled receptors that couple to the endogenous Gq protein of S2 cells (13-15). In this system, ligand binding to the receptor results in the activation of the phosphoinositide (PI) pathway: hydrolysis of PIP2 by phospholipase C into InsP3 and 4,5-diacylglycerol, and release of Ca2+ from intracellular stores. The stimulus-activated change in [Ca2+]i can be monitored with Ca2+-sensitive fluorescent ratiometric indicators, such as fura 2 (16).
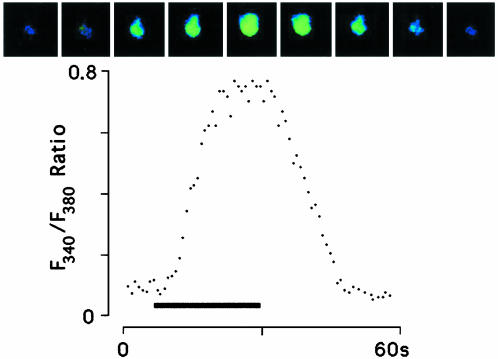
We transiently expressed Gr5a in S2 cells, loaded them with 100 μM fura 2, and applied 100 mM trehalose via puffer pipette (Fig. 2). Stimulation evoked Ca2+ release: cell response developed within ≈5 s of ligand application and reached a peak intensity within ≈15 s of application. Upon removal of the ligand, the level of intracellular calcium gradually returned to the baseline. These data provided initial evidence that Gr5a encodes a functional trehalose receptor when expressed in S2 cells. The results also suggested the possibility that Gr5a-encoded receptor protein couples to the endogenous phosphoinositide pathway. We then cotransfected S2 cells with Gr5a and promiscuous G proteins: Gα15,Gα16, or Gα15 and Gα16 together (17, 18). These G proteins are known to couple a wide variety of G protein-coupled receptors to intracellular Ca2+ release. There was no significant increase in the response intensity to trehalose stimulation compared with the S2-Gr5a cells, consistent with the possibility that Gr5a couples efficiently to Gq.
Fig. 2.
Time course of trehalose response in S2-Gr5a cells. (Upper) A series of images of a single fura 2-loaded S2-Gr5a cell, taken at 5-s intervals. The first image is taken 5 s before the application of 100 mM trehalose. (Lower) A quantitative representation of the response of the same cell. Bar indicates stimulus period.
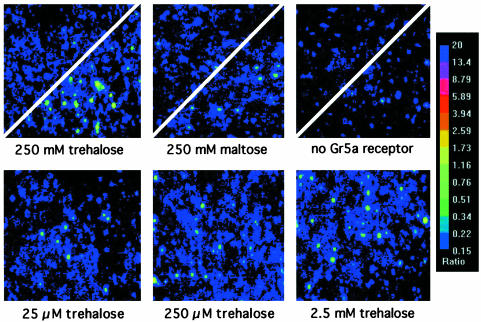
The response of S2-Gr5a cells depended on the presence of both an appropriate ligand and Gr5a (Fig. 3). We tested a wide range of trehalose concentrations, varying from 0.025 μM to 250 mM (Figs. 3 and 4a). Responses to trehalose show a steep dose-dependence over this range. Responses were weak at 25 μM, and saturation was observed in the low mM range (Fig. 4a). The Hill coefficient was nH = 1.92, suggesting the possibility that Gr5a functions as a homodimer.
Fig. 3.
Dose-dependence of trehalose response in S2-Gr5a cells. (Upper) Divided panels of S2-Gr5a cells (Left and Center) or negative controls, transfected with GFP vector alone (Right), before and after application of either 250 mM trehalose (Left and Right) or 250 mM maltose (Center). (Lower) Images of fields of S2-Gr5a cells taken on application of different concentrations of trehalose (indicated below).
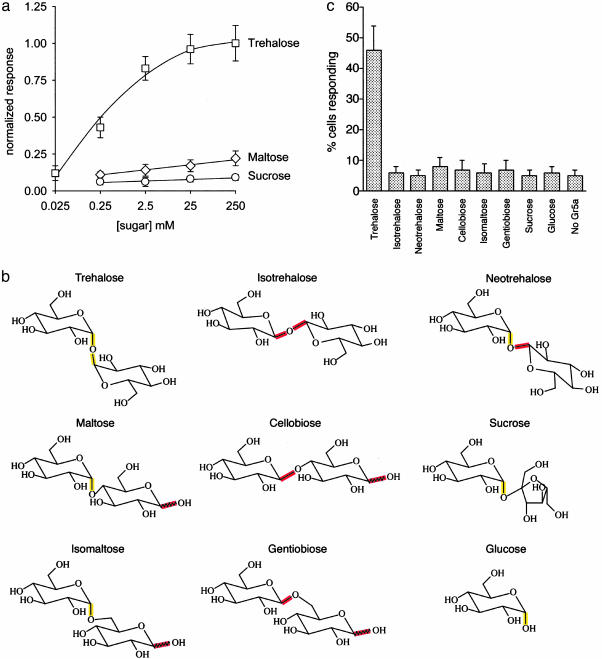
Fig. 4.
(a) Dose-response curves for selected disaccharides. Responses are normalized to response at 250 mM trehalose. 4 ≤ n ≤ 6; error bars = SEM. (b) Structures of selected disaccharides related to trehalose. With the exception of sucrose, all are composed of 2 units of glucose. The α-configuration is highlighted in yellow, and the β-configuration of the C1 carbon is highlighted in red. Isotrehalose and neotrehalose do not occur naturally. (c) Specificity of trehalose response in S2-Gr5a cells. Shown here are the responses of Gr5a-S2 cells to the various disaccharides illustrated in b. Compounds were tested at a concentration of 250 mM. 5 ≤ n ≤ 9. ”No Gr5a” cells were stimulated with 250 mM trehalose.
We have investigated the ligand specificity of Gr5a by challenging S2-Gr5a cells with other sugars. We first tested two other dissacharides, maltose (composed of two glucose units) and sucrose (one glucose unit linked to fructose), and found that even when tested at high concentrations they elicited little if any response, as measured in terms of the percentage of cells that yielded a F340/380 ≥ 0.34 (Figs. 3 and 4). We then systematically tested a number of common disaccharides structurally related to trehalose, each composed of two glucose units (Fig. 4b). These isomers varied only in the positions of their glycosidic bond (e.g., 1,1′, 1,4′, or 1,6′) and/or the configuration (α or β) of the d-glucose subunits. None of these other disaccharides evoked significant responses (Fig. 4c). The isomers tested included isotrehalose and neotrehalose, which, like trehalose, contain 1,1′ linkages; however, isotrehalose contains a β,β linkage, and neotrehalose contains an α,β linkage, whereas trehalose contains an α,α linkage. d-glucose, a monosaccharide component of all of the disaccharides tested, evoked no detectable Ca2+ release even at the highest dose tested (250 mM). The simplest interpretation of these results is that Gr5a recognizes moieties close to the 1α,1′α glycosidic bond.
Trehalose is found in certain drought-adapted organisms such as yeasts and is thought to play a role in protecting the membrane during dehydration (19, 20). Although trehalose has no effect on Ca2+ levels in control cells that do not express Gr5a, we carried out two further experiments to control for the formal possibility that trehalose interacts with the plasma membrane and somehow activates the receptor nonspecifically. We found first that trehalose had no effect on cells transfected with another Gr gene, Gr64f, and, second, that other molecules believed to have similar effects on membranes, glycerol and 1,2 propanediol, have no effect on Gr5a-expressing cells (tested at 100 mM each, data not shown). These results are consistent with the conclusion that trehalose is a ligand for Gr5a.
Discussion
This study provides direct evidence that Gr5a, a member of a large family of G protein-coupled receptors, functions as a trehalose taste receptor. Gr5a is expressed in neurons in taste sensilla of both the labellum and tarsi, consistent with a role as a taste receptor. When expressed in Drosophila S2 cells, it confers a response to trehalose in a dose-dependent manner. The response depends both on the expression of Gr5a and on a specific stimulus, trehalose. Other disaccharides tested evoke little if any response in this system.
We have provided evidence that Gr5a is expressed in all, or almost all, of the ≈33 sensilla present on the labellum. Because the labellum responds to a variety of sugars, and because the sensilla each contain a single sugar-sensitive neuron, the broad expression we have observed is consistent with a model in which many, if not all, of the sugar-sensitive taste neurons express more than one receptor. This model is supported by our earlier finding that mutation of Gr5a affected the physiological response of the sugar cell to trehalose, but not to sucrose, as if many of the sugar-sensitive cells contain both a trehalose receptor, Gr5a, and a sucrose receptor (9).
The expression pattern of Gr5a is broader than that observed for previously described GAL4 lines established by using promoters of other Gr genes (4, 5). The broad pattern is consistent with our earlier physiological data (9), which indicated that Gr5a is required for trehalose response in all L- and M-type sensilla (21). Hiroi et al. (22) also found that most sensilla on the labellum respond to trehalose.
We note that the response threshold of S2-Gr5a cells to trehalose is lower than in taste neurons in vivo, as determined in single-unit electrophysiological recordings (9). There are several possible explanations for this difference. One is that the two experiments measure different parameters, i.e., Ca2+ levels vs. action potential frequency, and the Ca2+ level we have established as a criterion for scoring a response may be less than that required to initiate action potentials. Another possibility concerns access to tastant: in the expression system, the cells are bathed in tastant, whereas in vivo the tastant must enter a pore in a sensillum and diffuse into the lymph surrounding a dendrite, where its final concentration may be lower than that of the test solution. A third possible explanation is that the density of receptor protein, or of another signaling component, may be greater in the heterologous expression system than in vivo.
The simplest interpretation of our results is that Gr5a functions as a homodimer, unlike the mammalian sweet receptors, which function as heterodimers (12). Furthermore, in contrast to the T1R2/T1R3 mammalian receptor that is rather broadly tuned to diverse sweet-tasting molecules such as sucrose, saccharin, dulcin, and acesulfame-K, the Gr5a receptor is tuned to trehalose and shows much less, if any, response to other sugars, such as sucrose, fructose, and glucose, which the fly encounters in its natural habitat.
The relatively narrow tuning of Gr5a has implications for the mechanism of taste coding. If other Drosophila taste receptors are as specific as Gr5a, then an individual tastant is likely to be encoded largely by the activity of one or a small number of receptors, as opposed to the integrated activity of many receptors, each exhibiting a varying degree of response to a ligand. In the olfactory system of Drosophila, many individual odorants activate several distinct classes of receptor neurons (23, 24), each expressing distinct odor receptors (ref. 25 and J.R.C., unpublished results). This model of taste coding is also supported by the severe loss of trehalose response after mutation of a single receptor gene, Gr5a. Analysis of further Gr proteins will be required to determine whether the narrow tuning of Gr5a is representative of Gr receptors at large or of those that recognize tastants of particular metabolic significance to the fly, such as trehalose.
Acknowledgments
We thank Dr. K. Morris and Mrs. M. Chyb for technical assistance and Dr. R. C. Hardie, Dr. S. Simon, Prof. J. L. Frazier, Dr. W. M. van der Goes van Naters, and A. Ray for discussion or comments on the manuscript. This work was supported by Biotechnology and Biological Sciences Research Council (S.C.), a National Research Service Award (to A.D.), and National Institutes of Health grants and a McKnight Investigator Award (to J.R.C.).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, ”Chemical Communication in a Post-Genomic World,” held January 17-19, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
References
- 1.Dethier, V. (1976) The Hungry Fly (Harvard Univ. Press, Cambridge, MA).
- 2.Rodrigues, V. & Siddiqi, O. (1978) Proc. Indian Acad. Sci. Sect. B 87, 147-160. [Google Scholar]
- 3.Clyne, P. J., Warr, C. G. & Carlson, J. R. (2000) Science 287, 1830-1834. [DOI] [PubMed] [Google Scholar]
- 4.Scott, K., Brady, R., Jr., Cravchik, A., Morozov, P., Rzhetsky, A., Zuker, C. & Axel, R. (2001) Cell 104, 661-673. [DOI] [PubMed] [Google Scholar]
- 5.Dunipace, L., Meister, S., McNealy, C. & Amrein, H. (2001) Curr. Biol. 11, 822-835. [DOI] [PubMed] [Google Scholar]
- 6.Chapman, R. F. (1982) in The Insects: Structure and Function (Harvard Univ. Press, Cambridge, MA), 3rd Ed., p. 919.
- 7.Tanimura, T., Isono, K., Takamura, T. & Shimada, I. (1982) J. Comp. Physiol. 147, 433-437. [Google Scholar]
- 8.Tanimura, T., Isono, K. & Yamamoto, M. (1988) Genetics 119, 399-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dahanukar, A., Foster, K., Van der Goes van Naters, W. M. & Carlson, J. R. (2001) Nat. Neurosci. 4, 1182-1186. [DOI] [PubMed] [Google Scholar]
- 10.Ueno, K., Ohta, M., Morita, H., Mikuni, Y., Nakajima, S., Yamamoto, K. & Isono, K. (2001) Curr. Biol. 11, 1451-1455. [DOI] [PubMed] [Google Scholar]
- 11.Bunch, T. A., Grinblat, Y. & Goldstein, L. S. B. (1988) Nucleic Acids Res. 16, 1043-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nelson, G., Hoon, M. A., Chandrashekhar, J., Zhang, Y., Ryba, N. J. P. & Zuker, C. S. (2001) Cell 106, 381-390. [DOI] [PubMed] [Google Scholar]
- 13.Hardie, R. C., Reuss, H., Lansdell, S. J. & Millar, N. S. (1997) Cell Calcium 21, 431-440. [DOI] [PubMed] [Google Scholar]
- 14.Chyb, S., Raghu, P. & Hardie, R. C. (1999) Nature 397, 255-259. [DOI] [PubMed] [Google Scholar]
- 15.Graziano, M. P., Broderick, D. J. & Tota, M. R. (1999) in Identification and Expression of G Protein-Coupled Receptors, ed. Lynch, K. L. (Wiley-Liss, New York), pp. 181-195.
- 16.Grynkiewicz, G., Poenie, M. & Tsien, R. Y. (1985) J. Biol. Chem. 260, 3440-3450. [PubMed] [Google Scholar]
- 17.Offermanns, S. & Simon, M. I. (1995) J. Biol. Chem. 270, 15175-15180. [DOI] [PubMed] [Google Scholar]
- 18.Milligan, G., Marshall, F. & Rees, S. (1996) Trends Pharmacol. Sci. 17, 235-237. [DOI] [PubMed] [Google Scholar]
- 19.Crowe, J. H., Crowe, L. M. & Chapman, D. (1984) Science 223, 701-703. [DOI] [PubMed] [Google Scholar]
- 20.Crowe, L. M. (2002) Comp. Biochem. Physiol. 131, 505-513. [DOI] [PubMed] [Google Scholar]
- 21.Ray, K., Hartenstein, V. & Rodrigues, V. (1993) Dev. Biol. 155, 26-37. [DOI] [PubMed] [Google Scholar]
- 22.Hiroi, M., Marion-Poll, F. & Tanimura, T. (2002) Zool. Sci. 19, 1009-1018. [DOI] [PubMed] [Google Scholar]
- 23.De Bruyne, M., Clyne, P. J. & Carlson, J. R. (1999) J. Neurosci. 19, 4520-4532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.De Bruyne, M., Foster, K. & Carlson, J. R. (2001) Neuron 30, 537-552. [DOI] [PubMed] [Google Scholar]
- 25.Dobritsa, A. A., Van der Goes van Naters, W., Warr, C. G., Steinbrecht, R. A. & Carlson, J. R. (2003) Neuron 37, 1-20. [DOI] [PubMed] [Google Scholar]