Abstract
The virulence of Pseudomonas syringae and many other proteobacterial pathogens is dependent on complex repertoires of effector proteins injected into host cells by type III secretion systems. The 28 well-expressed effector genes in the repertoire of the model pathogen P. syringae pv. tomato DC3000 were deleted to produce polymutant DC3000D28E. Growth of DC3000D28E in Nicotiana benthamiana was symptomless and 4 logs lower than that of DC3000ΔhopQ1-1, which causes disease in this model plant. DC3000D28E seemed functionally effectorless but otherwise WT in diagnostic phenotypes relevant to plant interactions (for example, ability to inject the AvrPto-Cya reporter into N. benthamiana). Various effector genes were integrated by homologous recombination into native loci or by a programmable or random in vivo assembly shuttle (PRIVAS) system into the exchangeable effector locus in the Hrp pathogenicity island of DC3000D28E. The latter method exploited dual adapters and recombination in yeast for efficient assembly of PCR products into programmed or random combinations of multiple effector genes. Native and PRIVAS-mediated integrations were combined to identify a minimal functional repertoire of eight effector genes that restored much of the virulence of DC3000ΔhopQ1-1 in N. benthamiana, revealing a hierarchy in effector function: AvrPtoB acts with priority in suppressing immunity, enabling other effectors to promote further growth (HopM1 and HopE1), chlorosis (HopG1), lesion formation (HopAM1-1), and near full growth and symptom production (AvrE, HopAA1-1, and/or HopN1 functioning synergistically with the previous effectors). DC3000D28E, the PRIVAS method, and minimal functional repertoires provide new resources for probing the plant immune system.
Keywords: effector-triggered immunity, pathogen-associated molecular pattern-triggered immunity, DNA shuffling, multigene recombineering, synthetic biology
Many proteobacterial pathogens of plants and animals disarm and infect their hosts by injecting 20–50 or more effector proteins through the type III secretion system (T3SS) (1). Studies focused on a few individual type III effectors (T3Es) in the repertoires of model pathogens have yielded seminal insights into host targets and T3E activities, but they also suggest that T3Es in a given repertoire, such as that of enteropathogenic Escherichia coli E2348/69, function in a “multifunctional, cooperative, and redundant” manner (2). That is, T3E repertoires may function as systems with properties beyond those of individual effectors.
The T3E repertoire of Pseudomonas syringae pv. tomato DC3000, which can cause disease in tomato and model plants Arabidopsis thaliana and Nicotiana benthamiana, is particularly amenable to systems-level study (3). The DC3000 T3Es, which are designated as Hrp outer protein (Hop) or avirulence (Avr) proteins, have been comprehensively identified, and 28 T3Es have been shown to be well-expressed and deployed during infection (4–7). The activities and targets in plants of several of these T3Es have been determined (8).
According to a current model for plant–pathogen interactions (9), the primary function of P. syringae T3Es is to suppress pathogen-associated molecular pattern (PAMP)-triggered immunity (PTI), which is elicited by common bacterial factors, such as flagellin, interacting with pattern recognition receptors (PRRs) on plant cell surfaces. Plants have a defense against T3Es that is based on detection inside plant cells of their presence or activity by resistance (R) proteins, which results in effector-triggered immunity (ETI). Pathogens may evade ETI by eliminating effectors that have become avirulence determinants or deploying other effectors that suppress ETI. This model predicts a coevolutionary process that would generate the observed amplification and polymorphism in genes encoding effectors in pathogens and PTI components and R proteins in plants. Determining general properties of these complex interaction systems, which also occur with many pathogenic fungi, oomycetes, and nematodes and their comparable effectors, has practical potential because of the widespread use of resistance breeding for crop protection and the frequent failure of resistance in the face of pathogen variation in the field (10).
The majority of the well-expressed DC3000 T3Es are encoded within six clusters in the DC3000 genome (11). Deletions of individual clusters revealed HopQ1-1 to function as the sole avirulence determinant for DC3000 in N. benthamiana, a plant that is particularly amenable to high-throughput genetic manipulation and bacterial growth assays (11, 12). Combinatorial deletions revealed only a small reduction in growth in N. benthamiana with the loss of 15 T3E genes in five clusters but a stronger reduction with the loss of just two or three T3E genes in either of two redundant effector groups (REGs). For example, a strong reduction in growth accompanied the combined loss of avrPto and avrPtoB, which comprises one REG. These observations suggest that the composition of T3E repertoires is functionally structured (13), but the difficulty of constructing alternative combinatorial polymutants has limited further exploration of interplay and redundancy in the DC3000 T3E repertoire.
We report the construction of a functionally effectorless derivative of DC3000, designated DC3000D28E (deficient in 28 effectors), development of a dual adapter recombination method for use with a programmable or random in vivo assembly shuttle (PRIVAS) system that enabled partial reassembly of the T3E repertoire in DC3000D28E, and identification of a minimal functional repertoire of T3Es that restores near WT growth and symptom production in N. benthamiana.
Results
Construction of P. syringae pv. tomato DC3000D28E, a Functionally Effectorless Polymutant.
We had previously constructed CUCPB5500, which lacks all 18 of the well-expressed T3E genes occurring in clusters (13). Here, we deleted the remaining 10 well-expressed T3E genes, again using pK18mobsacB (11), to produce polymutant DC3000D28E (CUCPB5585 in Fig. 1). Fig. S1 provides an overview of our genetic manipulations of DC3000 and depicts relevant genes and clusters as well as T3E pseudogenes and genes that seem to be only weakly expressed (4–7).
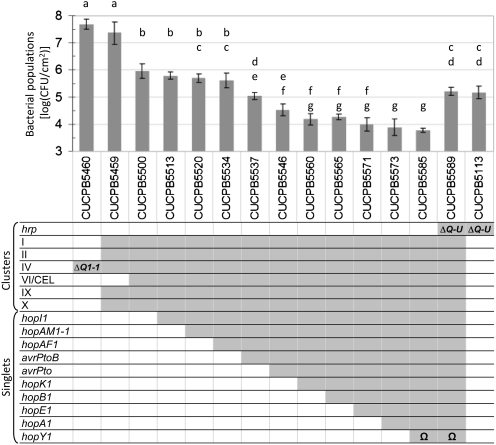
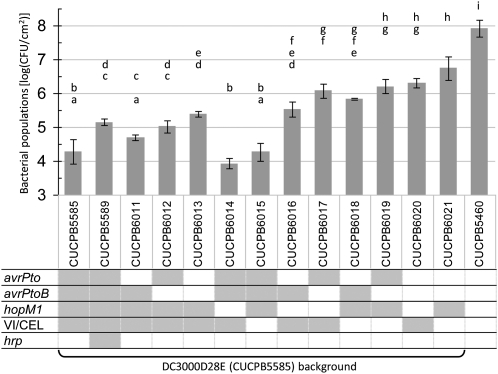
Fig. 1.
Deletion of 10 T3E genes in CUCPB5500 yields DC3000D28E (CUCPB5585), which is strongly reduced in growth in N. benthamiana. Bacteria were inoculated with a blunt syringe at 3 × 104 cfu/mL in MgCl2 buffer, and populations in three 0.8-cm leaf disks were determined at 6 dpi. The arithmetic mean of log-transformed values of four replicate infiltrations from independent plants is displayed ± SD. Strains with the same letter are not statistically different based on a Tukey's honestly significant difference (HSD) test (α = 0.05). The cells in the genotype grid are shadowed in dark gray if the corresponding strains carry a deletion of the matching genomic region or are left white if the locus is WT. The Ω for hopY1 indicates that the gene was interrupted by insertion of a spectinomycin resistance cassette flanked by FRT sites. This experiment was repeated three times with similar results.
Analysis of the Ability of DC3000D28E and Progenitors to Grow in N. benthamiana.
CUCPB5500, DC3000D28E, and intermediate polymutants with successive T3E gene deletions were analyzed for their ability to grow in N. benthamiana. Leaves were inoculated with test strains at 3 × 104 cfu/mL by infiltration with a blunt syringe and assayed 6 d postinoculation (dpi) for bacterial population levels (Fig. 1). Notably strong reductions in bacterial growth were observed with the successive deletions of avrPtoB and avrPto. The population levels of DC3000D28E were approximately 4 logs lower than that of DC3000ΔhopQ1-1 and lower than that of CUCPB5113, a T3SS-deficient DC3000 ΔhrcQb-hrcU (hrcQ-U) mutant. This observation prompted us to construct CUCPB5589, which is a ΔhrcQ-U derivative of DC3000D28E. Population levels of CUCPB5589 and CUCPB5113 were indistinguishable (Fig. 1). These observations suggested that DC3000D28E was functionally effectorless and revealed a potential for the DC3000 T3SS machinery to stimulate plant defenses.
Functional Analysis of DC3000D28E.
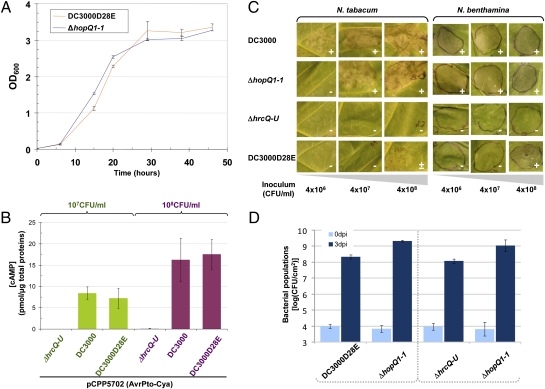
To determine whether DC3000D28E met key criteria for being functionally effectorless but otherwise WT in planta, we asked whether the mutant could grow robustly in apoplast-mimicking minimal media, deliver a translocation reporter into plant cells, be strongly reduced in its ability to elicit cell death in plants, and grow to high levels in planta in the presence of another strain that is able to defeat plant immunity. DC3000D28E grew similar to DC3000ΔhopQ1-1 in mannitol-glutamate minimal medium (14) (Fig. 2A) and Hrp minimal medium (Fig. S2B) but more slowly in rich Kings B medium (Fig. S2 A and C). DC3000D28E carrying pCPP5702, a plasmid expressing avrPto-cya from its native promoter (15), translocated the reporter as well as DC3000(pCPP5702) (with two levels of inoculum used to ensure that the assay was not saturated) (15) (Fig. 2B). DC3000D28E was compared with DC3000 and DC3000ΔhopQ1-1 for its ability to elicit cell death in N. benthamiana and nonhost N. tabacum at three inoculum levels chosen to exceed the threshold typically needed for elicitation of cell death associated with ETI. In both plants, 100 times more DC3000D28E was needed to elicit cell death 48 h after inoculation (Fig. 2C). Finally, DC3000D28E was compared with the DC3000 ΔhrcQ-U T3SS− mutant for its ability to grow in N. benthamiana when coinoculated with DC3000ΔhopQ1-1 and found to grow 4 logs better than without DC3000ΔhopQ1-1 (Fig. 1) and at least as well in this test as the ΔhrcQ-U mutant (Fig. 2D). Collectively, these observations suggest that DC3000D28E is functionally effectorless; although inexplicably growing more slowly in a rich medium, it does not seem to have second-site mutations that impair its ability to grow in planta and therefore, is suitable for testing T3Es for their ability to restore bacterial growth and induction of plant responses.
Fig. 2.
DC3000D28E (CUCPB5585) is functionally effectorless but otherwise WT in diagnostic assays. (A) Equivalent growth of DC3000D28E and DC3000ΔhopQ1-1 in liquid mannitol glutamate minimal medium supplemented with 50 μM iron citrate (means ± SD of the absorbance of triplicate cultures). (B) Equivalent translocation of an AvrPto-Cya fusion by DC3000D28E and DC3000. Plasmid pCPP5702 encoding the reporter gene under transcriptional control of the avrPto promoter was introduced into DC3000D28E, DC3000, and the ΔhrcQ-U T3SS− mutant. The resulting strains were infiltrated into N. benthamiana leaves at two different densities (107 and 108 cfu/mL) to control for assay saturation. cAMP concentrations were determined from tissues sampled from three independent leaves per treatment 7 h postinoculation. Means ± SD from one of two replicate experiments are shown. (C) Reduced ability of DC3000D28E to elicit ETI-like rapid plant cell death in N. benthamiana and N. tabacum. DC3000D28E and controls DC3000 (incompatible on both Nicotiana spp.), ΔhopQ1-1 (incompatible on N. tabacum), and ΔhrcQ-U cell suspensions in MgCl2 buffer, adjusted to three densities covering the dynamic range of the assay, were infiltrated into leaves, and the plant response was photographed 48 h later. Cell death response: +, positive; −, null; ±, partial. Each experiment was repeated at least three times with similar results. (D) Ability of DC3000D28E to be transcomplemented in mixed infections with virulent DC3000ΔhopQ1-1. Equal volumes of DC3000D28E and ΔhopQ1-1 strains (Left) or ΔhrcQ-U and ΔhopQ1-1 strains (Right) standardized at 3 × 106 cfu/mL (an inoculum level high enough for DC3000ΔhopQ1-1 to produce conditions favoring bacterial growth throughout the inoculated tissue) were mixed and infiltrated into leaves of N. benthamiana. Values (means ± SD of three samples) for the query strains and DC3000ΔhopQ1-1 were calculated by subtracting colony-forming unit counts on nonselective media (all strains) from colony-forming units on selective media (spectinomycin-resistant query strains). The experiment was repeated three times with similar results.
Recombination into Native Loci of Genes Representing Two REGs Reveals That AvrPto and AvrPtoB Act at an Early Phase of the Plant Immune Response.
Previous work highlighted the importance of the AvrPto/AvrPtoB and AvrE/HopM1/HopR1 REGs (13). We began the process of experimental reassembly of the DC3000 T3E repertoire by integrating avrPto, avrPtoB, hopM1, and the entire conserved effector locus (CEL or cluster VI, comprising avrE, hopM1, hopAA1-1, and hopN1) into their native locations in the genome of DC3000D28E by using pK18mobsacB. AvrPto and AvrPtoB both promoted significant growth, but neither HopM1 nor the entire set of CEL T3Es had this effect (Fig. 3). However, HopM1 was able to promote growth when combined with AvrPto or AvrPtoB, with maximal growth in these experiments occurring when AvrPtoB was combined with the complete CEL. Thus, AvrPto and AvrPtoB differ from members of the AvrE REG in seeming to be early-acting effectors. Weak additive effects on growth promotion were observed with T3Es from the same REG, but stronger effects were observed when representatives of two REGs were combined (for example, when AvrPtoB was combined with HopM1 rather than AvrPto). These observations suggest a hierarchy in the action of P. syringae T3Es, they provide a baseline involving natively restored T3E genes for analyzing subsequent experiments involving engineered assemblies of T3Es, and they reveal that just two T3Es can promote 2 of 4 logs of growth capacity that distinguish DC3000D28E and DC3000ΔhopQ1-1.
Fig. 3.
Restoration to native loci in DC3000D28E of genes encoding the AvrPto and AvrE REGs suggests that AvrPto and AvrPtoB interfere at an early phase of the host immune response. White fill in the genotype grid indicates that the locus was restored (note that the CEL includes hopM1). Strains harboring combinations of multiple WT loci were constructed by sequential integrations. Growth assays were performed at 6 dpi as in Fig. 1 with means ± SD of the bacterial populations calculated using values from three replicate leaves per strain. Means with the same letters are not statistically different based on a Tukey's HSD test (α = 0.05). This experiment was repeated three times with similar results.
Development of the PRIVAS System.
We exploited the ability of short terminal adapters to direct recombination of unrelated DNA fragments in yeast (16), and we developed a system of dual adapters to enable flexible assembly of multigene sets in shuttle vectors. Fig. 4 illustrates the application of the PRIVAS system to the assembly of T3E multigene sets and their subsequent integration into the naturally polymorphic exchangeable effector locus (EEL) of P. syringae (17). By choosing the desired flexible adapters from a premade panel of adapters and varying the composition of the pool of substrates for DNA assembly, the system can construct programmed or random gene sets. The shuttle vector replicates in yeast and E. coli and carries a 1,062-bp region of the DC3000 EEL. T3E genes used in this system are all expressed from native promoters and are cloned with their cognate chaperone genes where appropriate. To validate the PRIVAS system, we used it in programmed mode to introduce shcM-hopM1 into mutant CUCPB5515 (ΔIVΔCEL) and also avrPtoB and shcM-hopM1, sequentially, into DC3000D28E to reproduce the early-acting effector phenotype described in Fig. 3. In all cases, growth promotion was equivalent for T3E genes restored by PRIVAS and homologous recombination into native loci (Fig. S3).
Fig. 4.
The programmable or random in vivo assembly shuttle (PRIVAS) system exploits dual adapter recombination for facile integration of combinatorial gene sets into the DC3000 exchangeable effector locus (EEL). (A) Structure of a typical T3E genetic unit (GU) for PRIVAS. Primary PCR reactions with gene-specific oligonucleotide primers harboring 20-bp 3′ extensions amplify GUs flanked on each side by universal adaptor (UA) regions 1 or 2. (B) Secondary PCR reactions use these primary products as templates and flexible adapter (FA) primers composed of UA-specific segments at their 3′ end and one of a set of ~35-bp FA-homology regions at their 5′ end to yield UA-FA dual adapter-flanked GUs that are used as the elementary building blocks for in vivo assembly in yeast. The configuration of the gene sets, including gene orientation, can be fully programmed during construction by designing FA-flanked GUs so that a unique combination of recombination events between FAs leads to the closure of a circular DNA molecule containing the sequences of the shuttle vector as depicted in B for 3 GUs. The shuttle vector's backbone provides the origins of replication and selection markers for yeast and E. coli as well as an origin of transfer for conjugation into P. syringae. After transformation with a suitable pool of GUs and the linearized shuttle vector, plasmid DNA is extracted from yeast cells surviving selection and transferred into E. coli for subsequent conjugation into a recipient P. syringae strain and single cross-over integration into the EEL. (C) PRIVAS also can be used in random mode for the creation of complex combinatorial libraries of gene sets of variable configuration but of fixed size (equal to three in C). If several distinct GUs sharing the same pair of external FAs specifying a given position within the gene sets are included in the assembly reaction, identical FAs compete for recombination and hence, incorporation in growing DNA molecules. The final circular products contain polymorphic sets composed of GUs drawn from distinct bins of GUs at desired positions as illustrated in C.
Identification of Randomly Assembled T3E Gene Sets That Promote DC3000D28E Growth in N. benthamiana.
We then used the PRIVAS system in random mode to seek T3Es that could enhance bacterial growth of CUCPB6016 (DC3000D28E with avrPto and shcM-hopM1 restored to native loci). CUCPB6016 was chosen, because it contains representatives of the AvrPto and AvrE REGs, and the use of AvrPto allowed AvrPtoB, with its stronger growth phenotype, to function as a positive control in the randomized PRIVAS procedure. We chose 15 T3E-based genetic units (GUs) to form a manageable pool for initial construction of randomized T3E sets comprised of 3 or 5 GUs, with the shcF-hopF2-hopU1, hopAO1-shcV-hopV1, and shcO1-hopO1-1-hopT1-1 operons each comprising 1 GU. Seven of the DC3000 T3E genes were excluded, because they were associated with the CEL and the AvrE REG, whose growth contributions had been partially characterized (avrE, hopN1, hopAA1-1 and its paralog hopAA1-2, and hopR1), potentially inhibited bacterial growth (hopD1) (13), or seemed less common among sequenced P. syringae strains (hopB1) (8). CUCPB6016-PRIVAS strains were randomly chosen from the library for growth tests in N. benthamiana, where they were individually inoculated in four experiments involving 44 strains, each at ~3 × 104 cfu/mL, and compared with CUCPB6022 (CUCPB6016 empty integrated PRIVAS vector control) and CUCPB5459 (ΔIΔIIΔIVΔIXΔX) for growth at 6 dpi. The strains tested included 92 from the libraries with 3-GU sets and 84 with 5-GU sets. As shown for a representative experiment, the 44 strains in each batch produced a continuum of population levels (Fig. S4A). DNA sequencing was then used to identify the introduced T3E genes in 56 strains chosen to represent the extremes of the phenotypic continuum in the four batches and two classes of GU set size. This analysis yielded several key findings (Fig. S4B). avrPtoB was present in all of the strains with the strongest growth. hopE1 was also prevalent in these strains and in strains lacking avrPtoB that showed intermediate growth. Chlorosis was observed with some strains showing at least intermediate growth, and all of these strains contained hopG1. In summary, the PRIVAS procedure permits facile introduction of random small sets of T3Es into the genome of DC3000D28E, and it suggests that a few of the 18 T3Es present in the experimental pool may be particularly important in pathogenesis, although no combination restored virulence to near WT levels and alternative T3E gene combinations could produce relatively strong growth.
Identification of a Minimal Functional T3E Repertoire for Virulence in N. benthamiana.
We next used the PRIVAS system in programmable mode to introduce a series of T3E genes into CUCPB6017 (DC3000D28E with avrPtoB and shcM-hopM1 restored to native loci). We chose CUCPB6017, because the stronger growth contribution of AvrPtoB supported our goal of identifying a minimal functional repertoire. We chose three effector genes to add next, because observations with the random PRIVAS experiment or the deletion series leading to DC3000D28E indicated a role in growth (hopE1), chlorosis (hopG1), or lesion cell death (hopAM1). Indeed, introduction of hopE1 into CUCPB6017 produced a small but significant increase in growth (Fig. 5B). Introduction of hopE1+hopG1 or hopE1+hopAM1-1 produced chlorosis and some cell death, respectively, if the inoculum level was raised to 3 × 105 (Fig. 5A). Combining these five T3E genes yielded an increase in growth that was significantly above the level promoted by avrPtoB+hopM1+hopE1 but substantially below the level of DC3000ΔhopQ1-1 (Fig. 5B). Finally, we used PRIVAS to introduce hopE1+hopG1+hopAM1-1 into CUCPB6019 (DC3000D28E with avrPtoB and the complete CEL restored to native loci). The resulting strain, CUCPB6032, produced robust symptoms in N. benthamiana and achieved population levels that were more than 3 logs better than DC3000D28E and within a log of DC3000ΔhopQ1-1 (Fig. 5). Thus, a minimal set of eight T3Es is sufficient to restore the virulence of DC3000D28E in N. benthamiana to near WT levels.
Fig. 5.
Successive PRIVAS-mediated integration of eight T3Es into DC3000D28E reveals a hierarchy contributing to chlorosis, lesion formation, and near WT growth in N. benthamiana. PRIVAS was used in programmed mode to create various combinations of hopE1, hopAM1-1, and hopG1. The resulting gene sets were integrated at the EEL of DC3000D28E derivatives CUCPB6017 or CUCPB6019, which had avrPtoB and hopM1, respectively, or the entire CEL natively restored, as indicated by white-filled cells in the genotype grid. (A) Symptoms in N. benthamiana leaves. Leaves were infiltrated with two levels of inoculum, and the plants were kept in a chamber with 70–80% relative humidity. The fraction of plants showing symptoms and the nature of symptoms scored is shown; plus sign indicates chlorosis, and asterisk indicates cell death. (B) Bacterial growth in N. benthamiana. Bacteria were inoculated at 3 × 104 cfu/mL, and populations measured 6 dpi. The least-squares means ± SD of log (cfu/cm2) are shown. Means with the same letter are not significantly different using the Tukey–Kramer multiple comparisons method (α = 0.05). Eight independent experiments were performed with different subsets of the nine strains shown, with a minimum of three plants per strain in every experiment and a total of 213 data points. Randomized block design was used for data analysis using the statistical analysis program SAS.
Discussion
Plant pathogenic bacteria in the genera Pseudomonas, Xanthomonas, and Ralstonia deploy large T3E repertoires that have several systems-level properties regarding their contribution to virulence (8, 13, 18, 19): (i) T3Es collectively are essential, (ii) no single T3E is essential, (iii) some T3Es can be assigned to REGs that redundantly target distinct processes in plant defense, (iv) T3E repertoires can be highly variable, even among strains pathogenic on the same host, and (v) heterologous expression and delivery of effectors from other strains, or even from oomycetes (20), can increase the virulence of WT strains. Here, we have learned additional properties of the DC3000 T3E repertoire in the context of interactions with N. benthamiana: (i) no single T3E is sufficient for significant virulence, (ii) some T3Es seem to interfere with an early phase of the plant immune response (i.e., by disrupting PAMP perception) such that other T3Es make a contribution to virulence only in their presence, (iii) early-acting effectors also seem to suppress defenses elicited by the T3SS machinery, (iv) T3Es in small groups with reduced redundancy can be observed to contribute in a hierarchical fashion to growth and symptom production, and (v) a minimal functional repertoire seems to require several effectors and members of at least two REGs. Before considering these generalizations in the context of specific T3Es and known host targets, we must discuss the discovery path to the minimal functional repertoire.
Our search involved iterative introductions of 24 of 28 well-expressed DC3000 T3E genes (Fig. S1). Growth phenotypes observed during repertoire disassembly led us to the initial introduction of members of the AvrPto and AvrE REGs and then to the use of one member of each REG as the foundation for construction of a minimal functional repertoire. The PRIVAS system in random mode enabled us to search a large number of combinations of 18 T3Es not known to be associated with these REGs or the CEL (other than the AvrPtoB-positive control). No single T3E gene newly introduced at the random PRIVAS stage made a notably strong contribution to growth, but several different T3E combinations made modest contributions. We chose to explore hopE1, hopG1, and hopAM1, and the minimal functional repertoire that we defined seems to involve some synergy between these three T3Es and the CEL T3Es, as seen by comparing the relative growth of CUCPB6032 with that of CUCPB6019 and CUCPB6031 in Figs. 3 and 5. Thus, although AvrPtoB and the CEL T3Es are clearly important, they are insufficient for a minimal functional repertoire (and it is possible that not all four of the CEL T3Es are necessary). However, our data strongly suggest that DC3000 requires at least six T3Es to grow and cause disease in N. benthamiana, a model plant that is not a natural host for WT DC3000 and seems to be unusually susceptible to a variety of pathogens (although possessing fully functional PTI and ETI systems) (12).
DC3000D28E growth in N. benthamiana is symptomless and 4 logs lower than DC3000ΔhopQ1-1. DC3000D28E seems to elicit plant defenses that are T3SS-dependent and additional to basal PTI. In this regard, it is noteworthy that DC3000D28E has the WT complement of T3SS helper proteins (except HrpW1), which fall into the overlapping functional classes of harpins, translocators, and lytic transglycosylases, and several of these proteins can elicit plant defenses (15). T3Es in the minimal functional repertoire restore virulence to DC3000D28E in the following approximate hierarchy. AvrPtoB partially suppresses T3SS- and PAMP-triggered immunity. Other T3Es then promote further growth (HopM1 and HopE1), chlorosis (HopG1), lesion formation (HopAM1-1), and then near full growth and symptom production (AvrE, HopAA1-1, and/or HopN1 functioning synergistically with the previous effectors). We postulate that introducing more T3E genes would incrementally increase virulence and restore redundancy, with limits to repertoire size in field populations being imposed by interactions with coevolving host ETI systems.
Our limited knowledge of specific T3E functions is consistent with the hierarchy observed in the minimal repertoire. AvrPtoB inhibits PRR coreceptor complexes involved in initial perception of pathogens (21). HopM1 destabilizes a plant ADP ribosylation factor (ARF) guanine nucleotide exchange factor (GEF) protein involved in vesicle trafficking and likely important for plant deployment of defense factors (22). HopG1 is localized to plant mitochondria and elevates levels of reactive oxygen species (23). HopAM1 is thought to manipulate defense-related responses to the hormone abscisic acid and also causes cell death when expressed in yeast cells (24, 25). AvrE may mimic activated G proteins and thereby, functionally overlap with HopM1 in disrupting vesicle trafficking (26). HopAA1-1 elicits cell death when expressed in yeast and plant cells (25). HopN1 is a cysteine protease that can suppress ETI-associated cell death (27). How these few T3Es function together to form a minimal repertoire may be complex, because T3Es can have multiple domains and interfering activities. For example, AvrPtoB also possesses an E3 ubiquitin ligase domain that can suppress ETI (28), and HopM1, AvrE, and HopAA1-1 elicit ETI-like cell death in N. benthamiana when individually delivered by the nonpathogen P. fluorescens expressing cloned P. syringae T3SS genes (11). An advantage of the DC3000D28E PRIVAS system is that it provides strong phenotypes and facile tools for dissection of T3Es and their interplay in near native settings.
As explained above, it is possible that a minimal functional repertoire could have been assembled with T3Es other than HopE1, HopG1, and HopAM1. Indeed, the sequenced strains P. syringae pv. syringae B728a and P. syringae pv. tabaci 11528 also cause disease in N. benthamiana, but their genomes lack hopE1, hopG1, hopAM1, hopAA1-1, and hopN1 (29, 30). Clearly, P. syringae can defeat plants with alternative T3E repertoires. However, the bacteria do not seem able to do so with just one or two T3Es. This presumably is a result of redundancy and consequent robustness in plant PTI/ETI perception and signaling networks, as revealed by recent reports that mutations in multiple signaling components are needed to significantly compromise plant immunity and that exhaustive genetic screens revealed no essential PTI component signaling downstream of PRRs (31–35). In this regard, it is noteworthy that the PRIVAS system, modified for use with Agrobacterium tumefaciens-based vectors, could be used for random and programmed combinatorial expression and silencing of plant immunity genes. More broadly, the use of flexible dual adapters for recombination, as exemplified with the PRIVAS system, represents an addition to the growing suite of multigene recombineering tools (36) that is particularly suited for deconvoluting internal redundancy and exploring functional structure in complex biological systems.
In the case of plant–pathogen interactions, using pathogens with PRIVAS-derived minimal repertoires to defeat plant immunity provides a means to efficiently probe defenses at the systems level and complements studies based on plant genetics. By understanding how the PTI system fails, we may better breed plants with enhanced PTI system robustness. Similarly, by understanding how pathogens evolve rapidly adaptable T3E systems, we may deploy combinations of R genes that confer more durable ETI in the field. In summary, DC3000D28E, the PRIVAS system, and minimal functional repertoires provide resources for accelerated study of T3Es and plant immune systems.
Materials and Methods
Bacterial Strains, Culture Conditions, and Virulence Assays.
Bacterial strains and plasmids are described in SI Materials and Methods, Construction and Usage of the Vectors for Genomic Gene Replacement and Complementation and Table S1. Mutations and restorations of complementing genes to native loci were performed with the suicide-eviction vector pK18mobsacB and then confirmed by PCR (11). Primers for plasmid and mutant construction are given in Table S2. Culture conditions, plant virulence assays, and Cya reporter translocation assays have been previously described (13, 15).
PRIVAS.
Primers used in primary and secondary PCRs were synthesized by Integrated DNA Technologies. PCR reactions were performed with the high-fidelity PrimeSTAR HS DNA Polymerase from Takara Bio Inc. GU genomic contexts are provided in Fig. S5. For programmed assembly of clusters of sizes 1–3 or 5 GUs, a pool of flexible adapter (FA)-flanked GUs (~100 ng of each) obeying one of the paths presented in SI Materials and Methods, Dual Adapter Recombination and Programmable or Random In Vivo Assembly Shuttle System together with 75 ng linearized pCPP6218 plasmid were transformed into yeast strain MaV203 from Invitrogen using the standard lithium acetate/polyethylene glycol procedure, and the resulting recombinant circular shuttle vectors were selected over recircularized empty vector using cycloheximide counterselection. Plasmids purified from yeast were introduced into E. coli S17-1 strain and then conjugated into P. syringae (13). Similar to the procedure for the analysis of random clusters, the configurations of the programmed clusters assembled with PRIVAS were checked by P. syringae colony PCR amplification (Fig. S6). The proper nature and orientation of the GUs were verified by sequencing of the amplified DNA region immediately downstream of the FAs. For the creation of random cluster libraries, the composition of the pools of competing GUs in parallel assemblies followed a near optimal balanced incomplete block design where the positions within the clusters represent blocks, and the set of 15 tested GUs is equivalent to treatment levels. The PRIVAS system is further described in SI Materials and Methods, Dual Adapter Recombination and Programmable or Random In Vivo Assembly Shuttle System, Fig. S7, and Tables S3–S6.
Acknowledgments
We thank Kent Loeffler for photography and Shamil Sadigov from the Cornell Statistical Consulting Unit for help with data analysis. This work was supported by National Science Foundation Plant Genome Research Program Grant DBI-0605059.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1013031108/-/DCSupplemental.
References
- 1.Kenny B, Valdivia R. Host-microbe interactions: Bacteria. Curr Opin Microbiol. 2009;12:1–3. doi: 10.1016/j.mib.2009.01.002. [DOI] [PubMed] [Google Scholar]
- 2.Dean P, Kenny B. The effector repertoire of enteropathogenic E. coli: Ganging up on the host cell. Curr Opin Microbiol. 2009;12:101–109. doi: 10.1016/j.mib.2008.11.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buell CR, et al. The complete genome sequence of the Arabidopsis and tomato pathogen Pseudomonas syringae pv. tomato DC3000. Proc Natl Acad Sci USA. 2003;100:10181–10186. doi: 10.1073/pnas.1731982100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Chang JH, et al. A high-throughput, near saturating screen for type III effector genes from Pseudomonas syringae. Proc Natl Acad Sci USA. 2005;102:2549–2554. doi: 10.1073/pnas.0409660102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Vinatzer BA, Jelenska J, Greenberg JT. Bioinformatics correctly identifies many type III secretion substrates in the plant pathogen Pseudomonas syringae and the biocontrol isolate P. fluorescens SBW25. Mol Plant Microbe Interact. 2005;18:877–888. doi: 10.1094/MPMI-18-0877. [DOI] [PubMed] [Google Scholar]
- 6.Schechter LM, et al. Multiple approaches to a complete inventory of Pseudomonas syringae pv. tomato DC3000 type III secretion system effector proteins. Mol Plant Microbe Interact. 2006;19:1180–1192. doi: 10.1094/MPMI-19-1180. [DOI] [PubMed] [Google Scholar]
- 7.Lindeberg M, et al. Closing the circle on the discovery of genes encoding Hrp regulon members and type III secretion system effectors in the genomes of three model Pseudomonas syringae strains. Mol Plant Microbe Interact. 2006;19:1151–1158. doi: 10.1094/MPMI-19-1151. [DOI] [PubMed] [Google Scholar]
- 8.Cunnac S, Lindeberg M, Collmer A. Pseudomonas syringae type III secretion system effectors: Repertoires in search of functions. Curr Opin Microbiol. 2009;12:53–60. doi: 10.1016/j.mib.2008.12.003. [DOI] [PubMed] [Google Scholar]
- 9.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 10.Poland JA, Balint-Kurti PJ, Wisser RJ, Pratt RC, Nelson RJ. Shades of gray: The world of quantitative disease resistance. Trends Plant Sci. 2009;14:21–29. doi: 10.1016/j.tplants.2008.10.006. [DOI] [PubMed] [Google Scholar]
- 11.Wei C-F, et al. A Pseudomonas syringae pv. tomato DC3000 mutant lacking the type III effector HopQ1-1 is able to cause disease in the model plant Nicotiana benthamiana. Plant J. 2007;51:32–46. doi: 10.1111/j.1365-313X.2007.03126.x. [DOI] [PubMed] [Google Scholar]
- 12.Goodin MM, Zaitlin D, Naidu RA, Lommel SA. Nicotiana benthamiana: Its history and future as a model for plant-pathogen interactions. Mol Plant Microbe Interact. 2008;21:1015–1026. doi: 10.1094/MPMI-21-8-1015. [DOI] [PubMed] [Google Scholar]
- 13.Kvitko BH, et al. Deletions in the repertoire of Pseudomonas syringae pv. tomato DC3000 type III secretion effector genes reveal functional overlap among effectors. PLoS Pathog. 2009;5:e1000388. doi: 10.1371/journal.ppat.1000388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bronstein PA, et al. Global transcriptional responses of Pseudomonas syringae DC3000 to changes in iron bioavailability in vitro. BMC Microbiol. 2008;8:209. doi: 10.1186/1471-2180-8-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kvitko BH, Ramos AR, Morello JE, Oh H-S, Collmer A. Identification of harpins in Pseudomonas syringae pv. tomato DC3000, which are functionally similar to HrpK1 in promoting translocation of type III secretion system effectors. J Bacteriol. 2007;189:8059–8072. doi: 10.1128/JB.01146-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raymond CK, Pownder TA, Sexson SL. General method for plasmid construction using homologous recombination. Biotechniques. 1999;26:134–138. doi: 10.2144/99261rr02. [DOI] [PubMed] [Google Scholar]
- 17.Alfano JR, et al. The Pseudomonas syringae Hrp pathogenicity island has a tripartite mosaic structure composed of a cluster of type III secretion genes bounded by exchangeable effector and conserved effector loci that contribute to parasitic fitness and pathogenicity in plants. Proc Natl Acad Sci USA. 2000;97:4856–4861. doi: 10.1073/pnas.97.9.4856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kay S, Bonas U. How Xanthomonas type III effectors manipulate the host plant. Curr Opin Microbiol. 2009;12:37–43. doi: 10.1016/j.mib.2008.12.006. [DOI] [PubMed] [Google Scholar]
- 19.Poueymiro M, Genin S. Secreted proteins from Ralstonia solanacearum: A hundred tricks to kill a plant. Curr Opin Microbiol. 2009;12:44–52. doi: 10.1016/j.mib.2008.11.008. [DOI] [PubMed] [Google Scholar]
- 20.Sohn KH, Lei R, Nemri A, Jones JD. The downy mildew effector proteins ATR1 and ATR13 promote disease susceptibility in Arabidopsis thaliana. Plant Cell. 2007;19:4077–4090. doi: 10.1105/tpc.107.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Shan L, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nomura K, et al. A bacterial virulence protein suppresses host innate immunity to cause plant disease. Science. 2006;313:220–223. doi: 10.1126/science.1129523. [DOI] [PubMed] [Google Scholar]
- 23.Block A, et al. The Pseudomonas syringae type III effector HopG1 targets mitochondria, alters plant development and suppresses plant innate immunity. Cell Microbiol. 2010;12:318–330. doi: 10.1111/j.1462-5822.2009.01396.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Goel AK, et al. The Pseudomonas syringae type III effector HopAM1 enhances virulence on water-stressed plants. Mol Plant Microbe Interact. 2008;21:361–370. doi: 10.1094/MPMI-21-3-0361. [DOI] [PubMed] [Google Scholar]
- 25.Munkvold KR, Martin ME, Bronstein PA, Collmer A. A survey of the Pseudomonas syringae pv. tomato DC3000 type III secretion system effector repertoire reveals several effectors that are deleterious when expressed in Saccharomyces cerevisiae. Mol Plant Microbe Interact. 2008;21:490–502. doi: 10.1094/MPMI-21-4-0490. [DOI] [PubMed] [Google Scholar]
- 26.Ham JH, et al. Multiple activities of the plant pathogen type III effector proteins WtsE and AvrE require WxxxE motifs. Mol Plant Microbe Interact. 2009;22:703–712. doi: 10.1094/MPMI-22-6-0703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.López-Solanilla E, Bronstein PA, Schneider AR, Collmer A. HopPtoN is a Pseudomonas syringae Hrp (type III secretion system) cysteine protease effector that suppresses pathogen-induced necrosis associated with both compatible and incompatible plant interactions. Mol Microbiol. 2004;54:353–365. doi: 10.1111/j.1365-2958.2004.04285.x. [DOI] [PubMed] [Google Scholar]
- 28.Rosebrock TR, et al. A bacterial E3 ubiquitin ligase targets a host protein kinase to disrupt plant immunity. Nature. 2007;448:370–374. doi: 10.1038/nature05966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vinatzer BA, et al. The type III effector repertoire of Pseudomonas syringae pv. syringae B728a and its role in survival and disease on host and non-host plants. Mol Microbiol. 2006;62:26–44. doi: 10.1111/j.1365-2958.2006.05350.x. [DOI] [PubMed] [Google Scholar]
- 30.Studholme DJ, et al. A draft genome sequence and functional screen reveals the repertoire of type III secreted proteins of Pseudomonas syringae pathovar tabaci 11528. BMC Genomics. 2009;10:395. doi: 10.1186/1471-2164-10-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tsuda K, Sato M, Stoddard T, Glazebrook J, Katagiri F. Network properties of robust immunity in plants. PLoS Genet. 2009;5:e1000772. doi: 10.1371/journal.pgen.1000772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Boudsocq M, et al. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464:418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chakravarthy S, Velásquez AC, Ekengren SK, Collmer A, Martin GB. Identification of Nicotiana benthamiana genes involved in PAMP-triggered immunity. Mol Plant Microbe Interact. 2010;23:715–726. doi: 10.1094/MPMI-23-6-0715. [DOI] [PubMed] [Google Scholar]
- 34.Li J, et al. Specific ER quality control components required for biogenesis of the plant innate immune receptor EFR. Proc Natl Acad Sci USA. 2009;106:15973–15978. doi: 10.1073/pnas.0905532106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Saijo Y, et al. Receptor quality control in the endoplasmic reticulum for plant innate immunity. EMBO J. 2009;28:3439–3449. doi: 10.1038/emboj.2009.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bieniossek C, et al. Automated unrestricted multigene recombineering for multiprotein complex production. Nat Methods. 2009;6:447–450. doi: 10.1038/nmeth.1326. [DOI] [PubMed] [Google Scholar]