Abstract
The alternative pathway of complement is important in innate immunity, attacking not only microbes but all unprotected biological surfaces through powerful amplification. It is unresolved how host and nonhost surfaces are distinguished at the molecular level, but key components are domains 19–20 of the complement regulator factor H (FH), which interact with host (i.e., nonactivator surface glycosaminoglycans or sialic acids) and the C3d part of C3b. Our structure of the FH19–20:C3d complex at 2.3-Å resolution shows that FH19–20 has two distinct binding sites, FH19 and FH20, for C3b. We show simultaneous binding of FH19 to C3b and FH20 to nonactivator surface glycosaminoglycans, and we show that both of these interactions are necessary for full binding of FH to C3b on nonactivator surfaces (i.e., for target discrimination). We also show that C3d could replace glycosaminoglycan binding to FH20, thus providing a feedback control for preventing excess C3b deposition and complement amplification. This explains the molecular basis of atypical hemolytic uremic syndrome, where mutations on the binding interfaces between FH19–20 and C3d or between FH20 and glycosaminoglycans lead to complement attack against host surfaces.
Keywords: structure and function, X-ray crystallography, hemolysis, kidney diseases, human mutations
Previously unencountered microbes invading a human body must be rapidly recognized and eliminated. This is the function of innate immunity, which includes the alternative pathway (AP) of complement. AP components can attack targets with hydroxyl or amine groups (i.e., all biological surfaces). This is a powerful defense mechanism, because there is rapid amplification leading to efficient opsonization or target lysis by the membrane attack complex (MAC). The AP attack is, therefore, also potentially dangerous for the host if one's cells and acellular structures are not protected.
The AP activation is based on spontaneous hydrolysis of C3 in plasma leading to production of C3b, which then randomly attaches onto any surface hydroxyl or amine group through a reactive thioester located on the C3d part [i.e., thioester domain (TED)] of C3b. If these surface-attached C3b molecules are not quickly inactivated to iC3b and C3d, C3b deposition is rapidly amplified by a positive enzymatic feedback loop, leading to opsonophagocytosis and formation of the lytic membrane attack complex. On host surfaces, which are naturally nonactivators of the AP, efficient down-regulation of bound C3b occurs in three ways: factor I-mediated cleavage of C3b to inactive iC3b, acceleration of the decay of the preformed C3 convertases, or inhibition of factor B binding to C3b. Factor H (FH) is required for all these. It also down-regulates C3b deposition on noncellular surfaces, such as the heparan sulfate-rich glomerular basement membrane. FH is, thus, essential for restricting AP attack against host surfaces while allowing AP attack against foreign surfaces (i.e., for target discrimination) (1). A long-standing central question in complement research has been how does FH distinguish the two types? It is known that sialylation of erythrocyte surface makes the cells nonactivators by enhanced FH binding (2, 3), and glycosaminoglycans on endothelial cells participate in making the latter nonactivators in the same way (4).
FH is composed of 20 homologous domains. Domains 1–4 are essential for various regulatory activities such as cofactor and decay accelerating activity (5, 6), whereas domains 19 and 20 (FH19–20) are essential for target discrimination (7, 8) and bind the C3d part of C3b (9), glycosaminoglycans (10), and sialic acids (11). Mutations in FH19–20 lead to a severe systemic disease, atypical hemolytic uremic syndrome (aHUS). aHUS, which often leads to end-stage renal disease, is characterized by damage to erythrocytes, thrombocytes, and endothelial cells (12). The characteristic cell damage in aHUS occurs when the mutated FH is unable to efficiently recognize C3b on these cells. The molecular mechanism of AP target discrimination is a major unresolved question, because some mutations interfere with binding of FH19–20 to cell surfaces and others interfere with C3b/C3d binding (13–15); the mutations are spread over both domains 19 and 20.
We have determined the X-ray crystal structure of FH19–20:C3d complex and performed extensive mutagenesis, binding, and functional studies. Results of these indicate how only two complement proteins, C3b and FH, can perform efficient target recognition. aHUS mutations in FH19, FH20, or two different parts of C3d all lead to cell destruction, because FH19–20 has two binding sites for C3d and C3d has two binding sites for FH19–20. In our comprehensive model of target discrimination, FH20 binds to glycosaminoglycans on host surfaces and FH19 binds to C3b, thus leading to cofactor activity. In addition, FH20 can also bind to any already deposited C3d on the surface, again allowing FH19 to bind another C3b and leading to its inactivation.
Results
Structure of the FH19–20:C3d Complex.
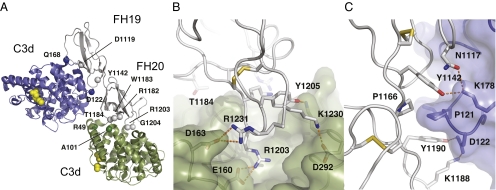
We expressed and purified C3d and FH19–20 and crystallized the complex (Materials and Methods). The structure was initially determined by molecular replacement with C3d at 3.8 Å but refined against a higher-resolution dataset at 2.3 Å (Table S1 and Fig. S1). The asymmetric unit reveals a surprising FH19–20:C3d2 heterotrimer, where two sites on a single FH19–20 bind two separate C3ds in different ways and at different sites (Fig. 1A). The overall structures of both C3d molecules are unchanged (16), whereas there are small local conformational changes in FH19–20 (13) because of binding. Both binding sites on C3d are far from the thioester site used for covalent attachment of C3b to targets (16) (Fig. 1).
Fig. 1.
Structure of the FH19–20:C3d complex. (A) The asymmetric unit contains one FH19–20 (gray) and two C3d molecules (green and blue). The residues mutated in aHUS patients and located at the interfaces are annotated and shown as green/blue spheres (C3d) or gray spheres (FH19–20). The thioester site residues are in yellow. (B) Close-up view of the FH20–C3d interface and (C) the FH19–C3d interface: FH19–20 is in gray, C3d is in green or blue, the main interface residues are sticks, disulphide bridges are in yellow, and hydrogen bonds are dashed lines. Structure figures were prepared using PyMol (version 1.3; Schrödinger, LLC).
Of the two interfaces on FH that bind to C3d, one interface (the FH20 site) is formed by the tip of the domain 20, which binds in a strongly electrostatic manner in a negatively charged cleft on the concave top of the α/α-helical bundle of C3d (the C3dFH20 site) (Fig. 1B and Fig. S2 A and C). The side chain of Arg1203 on FH20 protrudes into the cleft and forms an ion pair with C3d Glu160 (Glu1153 in the full sequence of C3). The total buried surface area of 490 Å2 includes three ion pairs and an extensive ionic network (Fig. S2 B and D), which presumably compensates for the low shape complementarity (Sc) (17) of 0.43, consistent with the observed micromolar affinities. There are local changes in the loops around the FH20 site: Lys1188 turns almost 180° to point to C3d, and Thr1184 and Arg1171 pack against C3d. The other interface (the FH19 site) is larger (620 Å2) and less charged (Fig. 1C and Fig. S2B) but with a higher Sc (0.70). This interface is formed by residues from both FH19 and FH20 that bind to the side of C3d at helices 104–118 and 170–189 and the Pro121 loop (the C3dFH19 site) (Fig. 1C and Fig. S2D).
The most important and striking feature of this complex is that the two sites are discontinuous and do not allow one FH to bind simultaneously one C3d through both interfaces. In addition, the C3dFH20 site is partially buried in the full C3b molecule (see below).
aHUS-Associated Mutations Are Found in Both Interfaces.
The unexpected structure with two binding sites explains why the 15 reported aHUS-associated point mutations in surface residues occur discontinuously in both FH domains 19 and 20 (Fig. 1A and Table 1). Of these, our structure suggests that 2 of 3 mutations in FH19 (Asp1119Gly and Tyr1142Asp) and 5 of 12 mutations in FH20 (Arg1182Ser, Trp1183Arg/Leu, Thr1184Arg, Arg1203Trp, and Gly1204Glu) would lead to impaired binding of FH19–20 to C3d/C3b. Five of the other eight residues affect binding of FH19–20 to heparin and/or endothelial cells (13–15) (Table 1 and Table S2). The remaining two mutations (Glu1135Asp and Ile1170Val) are partially buried and presumably affect FH folding.
Table 1.
Location and reported functional consequences of surface-exposed aHUS-associated mutations within FH19–20 and C3d part of C3b
| Reported functional defects* |
|||
| Location in FH19–20:C3d | Mutations | FH–C3b/d | FH–heparin |
| aHUS mutations on FH19–20 surface | |||
| FH19 site | Asp1119Gly† | ↓ | ↔ |
| FH19 site | Tyr1142Asp/Cys† | nt | nt |
| FH20 site | Arg1182Ser† | ↓ | ↓ |
| FH20 site | Trp1183Leu† | ↓ | ↓ |
| FH20 site | Thr1184Arg† | ↓ | ↑ |
| FH20 site | Arg1203Trp† | ↓ | ↓ |
| FH20 site | Gly1204Glu† | nt | nt |
| Heparin site? | Gly1194Asp† | nt | nt |
| Heparin site | Arg1206Cys | ↓ | ↓ |
| Heparin site | Arg1210Cys | ↓ | ↓ |
| Heparin site | Arg1215Gln/Gly | ↓ | ↓ |
| Next to heparin site? | Leu1189Arg | ↔ | ↑ |
| Next to heparin site? | Glu1198Ala | ↔ | ↑ |
| aHUS mutations on C3d surface | |||
| C3dFH19 | Asp122Asn† | ↓ | |
| C3dFH19 | Gln168Lys† | ↓ | |
| C3dFH20 | Arg49Leu† | nt | |
| C3dFH20 | Ala101Val† | ↓ | |
| Next to C3dFH19 | Ile164Thr† | nt | |
*Details of functional analyses and references are found in Tables S2 and S3. ↓, binding diminished because of the mutation; ↔, no effect; ↑, enhanced binding; nt, not tested.
†Mutants discussed in this report. Buried mutations likely to disrupt the fold are not listed: Val1134Gly, Glu1135Asp, Trp1157Arg, Cys1163Trp, Val1168Glu, Ile1169Leu, Ile1170Val, Ser1191Leu/Trp, Val1197Ala, Phe1199Ser, and Pro1226Ser for FH19–20 and Cys165Trp for the C3d part of C3b.
Some aHUS patients have mutations in C3. All five published mutations within the C3d domain are located either in or near C3dFH19 or C3dFH20 sites. Two mutations (Asp122Asn and Gln168Lys) affect residues in contact with the FH19 binding site, one mutation (Ile164Thr) is near that site, and two mutations (Arg49Leu and Ala101Val) affect residues in contact with the FH20 binding site on C3d (Table 1 and Table S3). Three of these mutations (Asp122Asn, Gln168Lys, and Ala101Val) are known to impair FH binding (18), whereas two have yet to be studied (19).
Verification of the Two FH-C3d Interfaces by Mutagenesis.
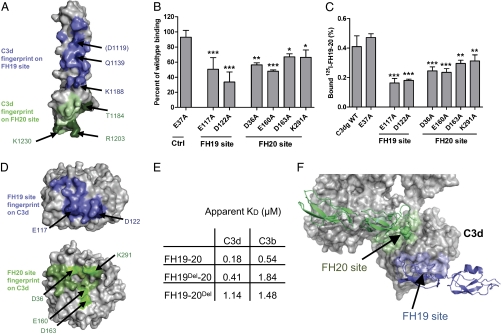
The presence of aHUS mutations on both sides of the FH19:C3d and FH20:C3d interfaces strongly suggests that neither interface is an artifact. To prove this, we analyzed published data and studied further mutations in both FH19–20 and C3d. All of the published point mutations in the FH19 (Asp1119Gly, Gln1139Ala, and Lys1188Ala) or FH20 site (Arg1182Ser/Ala, Trp1183Leu, Thr1184Arg, and Arg1203Ala) led to reduced C3d/C3b binding (13–15, 20) (Fig. 2A and Table 1). Next, we tested binding of seven C3d mutants to FH19–20. Both the mutations in the C3dFH19 site (Glu117Ala and Asp122Ala) and all four mutations in the C3dFH20 site (Asp36Ala, Glu160Ala, Asp163Ala, and Lys291Ala) significantly reduced the affinity of the interaction (Fig. 2 B–D), whereas the control mutation (Glu37Ala) had no effect.
Fig. 2.
FH19–20 binding sites on C3d and C3b. (A) Mutations on FH19–20 that reduce affinity to C3d or C3b (13–15, 20) are in darker color and annotated. (B) Binding of C3dg mutants to solid-phase FH19–20 by surface plasmon resonance (SPR) normalized to WT C3dg. Bars indicate SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001. (C) Binding of 125I-labeled FH19–20 to microtitre plate-coated C3dg mutants (SD indicated; n = 3) and (D) C3d mutations that impaired FH19–20 binding (darker blue and bright green). (E) Apparent affinities of FH19–20, FH19Del–20, and FH19–20Del to C3d and C3b measured using SPR. (F) Location of the exposed FH19 and partially occluded FH20 binding sites in C3b. FH19–20 bound to the FH20 binding site is shown in green, FH19–20 bound to the FH19 binding site is in blue, and C3b is in gray.
Availability of the Binding Sites on C3b.
We then generated multiple mutations in FH19–20 to delete either the FH19 or FH20 site, FH19Del-20 (Gln1137Ala-Gln1139Ala-Tyr1142Ala) and FH19–20Del (Thr1184Gly-Lys1202Ala-Arg1203Ala-Tyr1205Ala), and tested their binding to C3d and C3b. C3d bound WT FH19–20 with a twofold higher affinity than FH19Del-20 and a sixfold higher affinity than FH19–20Del (Fig. 2E). This indicates that the FH19 and FH20 sites bind C3d independently and with similar affinities. FH19–20Del bound similarly to C3d and C3b (apparent KD = 1.14 vs. 1.48 μM), indicating that the C3bFH19 site is fully available, as expected from the X-ray structure of C3b (21). Conversely, C3b bound WT FH19–20 or FH19Del–20 with a three to four times lower affinity than C3d did. This, although unanticipated based on prior information, is also consistent with the C3b structure (21), because the C3bFH20 site is partially hidden (Fig. 2F). Thus, conformational change in C3b would be required for full binding through the C3b:FH20 interface (Fig. S3). Furthermore, the C3dFH20 site also overlaps with the binding site of FH4 in the FH1–4:C3b structure (22). Despite this, we were unable to inhibit the cofactor or decay-accelerating functions of FH1–4 or full FH by either FH19–20 or FH19Del–20 (Fig. S4), indicating that FH4 is not essential for FH1–4 functions in the fluid phase, which is also consistent with earlier studies (5). Overall, it seems that FH19–20 binds C3d primarily through the FH20 site (Fig. 2E) (apparent KD = 0.41 vs. 1.84 μM) but binds C3b through the FH19 site. This has important implications for understanding how FH functions in target discrimination (see below).
Role of the Two Interfaces in Target Discrimination.
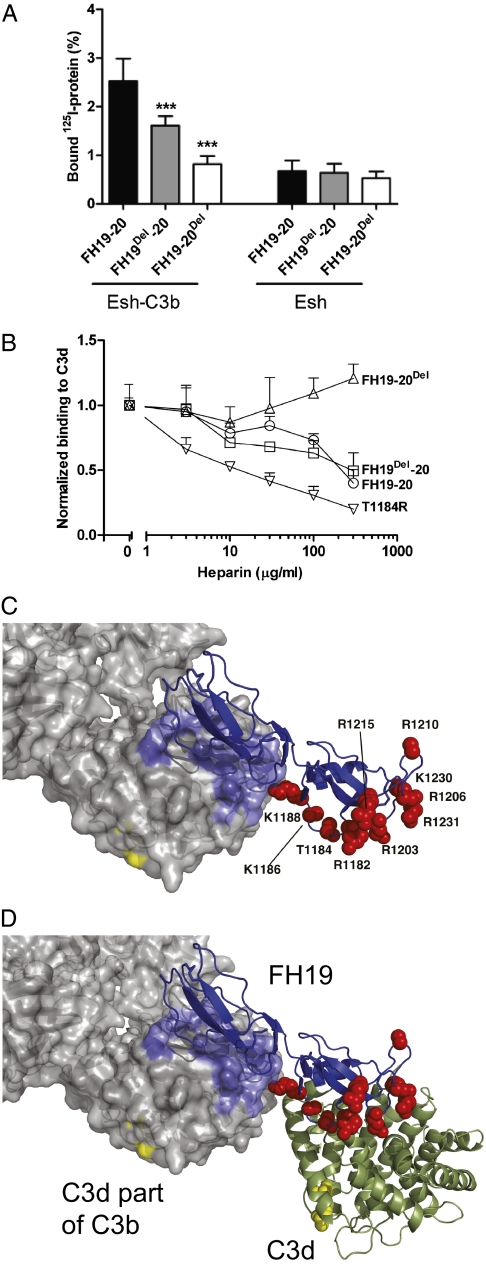
Next, we tested if both FH19 and FH20 sites are involved in recognizing C3b on a model nonactivator sheep erythrocyte surface (Esh). These cells are well-described nonactivator cells for human AP caused by the sialylated surface molecules (2, 3). Compared with WT FH19–20, binding of FH19Del–20 to Esh was reduced by 30–40%, whereas binding of FH19–20Del was practically abolished (Fig. 3A). Furthermore, heparin decasaccharide efficiently competed with FH19Del–20 but not FH19–20Del for C3d (Fig. 3B). This was as expected, because heparin binds to FH20 (10, 23) and interferes with C3d binding (24).
Fig. 3.
Role of FH19–20 interfaces in AP target discrimination. (A) Binding of FH19–20, FH19Del–20, and FH19–20Del to nonactivator particles [sheep erythrocytes (Esh)] with or without C3b depositions. Bars indicate SD (n = 3). ***P < 0.001. (B) Effect of heparin decasaccharide on binding of FH19–20 and its mutants to C3d. (C) Superimposition of C3b (21) and the FH19–20:C3d complex bound through the FH19 site showing availability of the heparin-binding residues (red spheres). (D) As in C, no steric clashes occur if another C3d simultaneously binds to the FH20 site. The thioester residues are in yellow, showing that this model implies that C3b and C3d are attached in the same way to the cell surface.
Discussion
The results above allow us to propose a comprehensive explanation for target discrimination by AP (i.e., down-regulation of complement amplification on host surfaces only), the molecular mechanism of aHUS associated with FH19–20 and C3b/C3d mutations, and the reason why most of the aHUS mutations are dominantly negative mutations.
The simplest model for target discrimination would be that FH20 recognizes and binds to cell surface polyanions such as glycosaminoglycans or putatively to sialic acids, while simultaneously, FH19 binds to the C3d part of C3b (Fig. 3C). The bidentate binding of C3b by FH19 and FH1–4 (22) (Fig. S3C) obviously would enhance the FH avidity and thus, its activity. Interestingly, heparin inhibited binding of C3d to the aHUS mutant FH19–20T1184R even more than to FH19Del–20 (Fig. 3B), suggesting that the abnormally strong heparin binding by this mutant (14) could interfere with C3d binding by the FH19 site, thus leading to aHUS.
However, if FH19 is the primary site for binding C3b, why does the FH20 site bind to C3d, and why are there aHUS mutations in the FH20 interface in both FH and C3b? There are two possibilities. A conformational change on binding of C3b to a nonactivator surface might expose the FH20 binding site. Although a conformational change in C3b on surface binding has been suggested (25), this seems unlikely to explain AP target discrimination. First, the overlap in the FH20 site for C3b and the glycosaminoglycan site on FH20 means that binding through FH20 would prevent binding to surface glycosaminoglycans. Second, such a conformational change would destroy the FH4 binding site on C3b.
The other, hitherto unrecognized possibility is that while FH19 binds to C3b, FH20 can bind an additional C3d, consistent with the fourfold higher affinity of the FH20 site for C3d than C3b (Fig. 2E). Our FH19–20:C3d2 structure shows that, although binding to two C3bs is sterically impossible, FH19–20 can simultaneously bind to C3b and C3d (Fig. 3D). It is also likely, although not tested, that the FH20 site could bind not only to C3d but also to iC3b, because EM studies suggest that the C3c part of iC3b moves away from C3d (26); this likely removes the partial obstruction of the FH20 site. In vivo, C3b is efficiently cleaved to iC3b and C3d on nonactivator cells but not on activator surfaces. C3d and iC3b, like polyanions, are in vivo markers of nonactivator surfaces and would, therefore, promote the shutdown of AP activation and amplification. Intriguingly, the two thioester sites on C3d and C3b both face to the putative cell surface in our C3b:FH19–FH20:C3d model structure—completely consistent with C3d (or iC3b) recruiting FH on nonactivator surfaces like we propose. Binding of the FH20 site to C3d or iC3b could, thus, replace binding of FH20 to nonactivator surface polyanions, further explaining why the C3d and polyanion sites overlap on FH20. The spatial organization of these complexes is similar (Fig. 3 C and D).
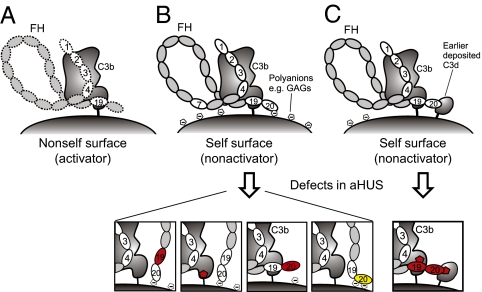
Our results suggest a comprehensive molecular mechanism for target discrimination of AP (Fig. 4) and provide evidence for negative feedback control of activation on nonactivator surfaces. On an activator (nonhost) surface, the carboxyl terminus of FH binds C3b monovalently and weakly through either the FH19 binding site or the partially occluded FH20 binding site on C3b, leading to relatively weak avidity of FH for C3b, poor control of C3b or C3bBb by FH1–4, subsequent AP activation, and elimination of the target. On nonactivator (host) surfaces, a dual contact of FH19–20 occurs: binding to C3b through the FH19 site and to cell surface polyanions through the FH20 glycosaminoglycan binding site. This increases the avidity of FH for nonactivator surface-bound C3b, leading to FH1–4 binding and rapid down-regulation of AP. This also implies negative feedback control activity. Breakdown of C3b to C3d (or iC3b) on nonactivator surfaces will increase the number of binding sites for FH19–20. More FH can be recruited, leading to a more rapid breakdown of C3b to iC3b and C3d. This also suggests why FH mutants can be dominantly negative; any mutant that prevents binding or slows down this regulatory feedback loop would lead to disease.
Fig. 4.
Model of regulation of complement amplification and its implications to pathogenesis of aHUS. (A) Weak binding on nonhost cells leads to activation. (B) In host cells, strong binding to glycosaminoglycans and C3b leads to regulation, except in mutated states (Lower). (C) In host cells, strong binding to previously deposited C3d (or possibly iC3b) and C3b also leads to down-regulation, except in mutated states in aHUS (Lower). Red and yellow denote loss- and gain-of-function mutations, respectively.
This model explains all published data on aHUS point mutations in FH19–20 and the C3d part of C3b (Fig. 4). Mutations in FH19 affect C3b binding (Fig. 2A), and mutations in FH20 affect, variously, glycosaminoglycan binding and C3d binding (the effect of the Gly1194Asp mutation is speculative, because it has not been experimentally validated) (Figs. 2A and 3B and Table S1). All mutations in the C3d domain affect either FH19 or FH20 binding (Fig. 2 B–D). Our model explains how all these mutations reduce control of C3b on nonactivator surfaces, leading to aHUS (Fig. 4). The model suggests intriguing future directions for study of biocompatibility of plasma-exposed materials and complement evasion by several FH19–20 binding pathogenic microbes. It also provides a platform for designing therapies for alternative pathway-associated diseases such as aHUS.
Materials and Methods
Proteins and Site-Directed Mutagenesis.
Mutagenesis, expression, and purification have been previously described for FH19–20 (27), most FH19–20 mutants (13), and C3d (or C3dg) and C3dg mutants (28). FH constructs were expressed in Pichia pastoris and C3d or C3dg constructs in Escherichia coli. New point mutations were introduced into the FH19–20 similarly to the previously described mutants using the QuikChange technique (Stratagene) (14). The template for generating the FH19Del-20 mutant was the Gln1139Ala mutant, and the primer used to generate the triple mutant was CAT CAG TTG AGT ACG CAT GCG CGA ACT TGG CTC AAC TTG AGG G. The template for FH19–20Del was the Arg1203Ala mutant, and the primers used to generate the quadruple mutant were CAG TTG AAT TTG TGT GTG CAG CGG GAG CTC GTC TTT CAT CAC G and CAT AGC ATT AAG GTG GGG AGC CAA ACA GAA GCT TTA TTC G. The FH19–20 mutants were expressed in P. pastoris (14), and FH19Del–20 was purified using heparin affinity chromatography; however, FH19–20Del was purified with the Resource S cation exchange column (GE Healthcare) because of the lack of heparin binding. C3 and FH were purified from plasma, and C3b was prepared from C3 as described (9, 29).
A fluorescence thermal shift assay to confirm mutant stability relative to WT was performed by adding 2.5 μL of 1:100 diluted Sypro Orange (Molecular Probes), 2.5 μL of 0.7 mg/mL protein, 20 μL of 0.1 M Hepes, and 20 μL of 0.15 M NaCl, pH 7.5, to wells of a 48-well thin-wall PCR plate (Bio-Rad). The plates were sealed with Optical-Quality Sealing Tape (Bio-Rad) and heated in MiniOpticon RT-PCR equipment (Bio-Rad) from 20 °C to 95 °C in increments of 1 °C. The wavelengths for excitation and emission were 490 and 575 nm, respectively. Tm values for FH19–20Del and FH19Del–20 decreased by 4 °C and 6 °C, respectively, compared with WT FH19–20 (66 °C).
Crystallization and Solution of the Structure.
Despite extensive trials, we were unable to crystallize FH19–20 with C3d, because FH19–20 always crystallized by itself as a tetramer (13, 20) in various crystal forms. We reasoned that inhibiting tetramer formation would help crystallization of the complex, and therefore, we used the FH19–20 double mutant Asp1119Gly–Gln1139Ala designed to break the tetramer interface (13). The mutant still bound C3d well but crystallized poorly by itself. This allowed us to crystallize FH19–20D1119G–Q1139A:C3d complex because the homotetrameric crystals did not form. Small-angle X-ray scattering (SAXS) data also showed that the FH19–20D1119G–Q1139A mutant is clearly monomeric, whereas the WT protein has a tendency to form higher oligomers and aggregates.
The FH19–20D1119G–Q1139A:C3d complex was crystallized at 22 °C from 0.1 M Hepes, pH 7.5, containing 14% PEG 4000 (160 μM FH19–20, 200 μM C3d). Crystals appeared in 3–4 d. All data were collected at the European Synchrotron Radiation Facility (ESRF). The initial dataset, to 3.8-Å resolution, was collected at ID14-2; optimization of cryocooling conditions with 10% glycerol enabled us to collect data to 2.3-Å resolution at ID23-1 (Table S1). The initial molecular replacement solution was found with the 3.8-Å data. PHASER (30) readily found two molecules of C3d but not FH19–20. However, closer inspection of Fo–Fc and σA-weighted maps revealed very clear features of continuous extra density consistent with a single FH19–20, and therefore, we manually built FH20 into the density between the two C3d molecules, giving an asymmetric unit with two C3d molecules and a single FH19–20. Successive rounds of building with Coot (31) and refinement with REFMAC (32) or PHENIX (33) against the 2.3-Å dataset allowed us to build an essentially complete model of the full heterotrimeric complex. The R factors (Rwork/Rfree) are 20.4%/24.3% with good geometry (Table S1). The C3ds are essentially unchanged from earlier structures [root mean square deviations (rmsds)/Cα = 0.27–0.5 Å], whereas there are local conformational changes in the FH (rmsd/Cα = 1.04 Å). Modeling of the two mutated side chains D1119 and Q1139 to the crystal structure (Fig. S5) indicated that Gln1139 would hydrogen bond to the Ile115 carbonyl group and Ser171, whereas Asp1119 would form an intermolecular helix cap with C3d (Fig. S5). The mutant should bind C3d less well than WT, which was observed.
Surface Plasmon Resonance Analyses.
The binding of C3dg mutants to FH19–20 was analyzed on a Biacore 3000 instrument (Biacore/GE Healthcare) essentially as described earlier using buffer (pH 7.2) containing 10 mM Hepes, 0.15 M NaCl, 3 mM EDTA, and 0.005% P20 surfactant (28). WT FH19–20 was coupled to CM-5 sensor chips to achieve immobilized protein level of ∼1,900 resonance units (RU). The concentration of the C3dg mutants in the fluid phase was 2.5 μM.
The surface plasmon resonance (SPR) analyses for testing affinity of the FH19–20 constructs to C3d and C3b were performed on a Biacore 2000 instrument using CM-5 sensor chips coupled with 500 RU C3d or 2,300 RU C3b to obtain an equal number of coupled molecules. Kinetic analyses were performed at 22 °C using PBS as the running buffer at 30 μL/min flow and 1.25–40 μM FH19–20 or its mutants.
Radioligand Assays.
To test binding of C3dg mutants to FH19–20, 80 μL C3dg or C3dg mutant (10 μg/mL) were coupled overnight onto Nunc Polysorp BreakApart wells, and the binding of 125I-FH19–20 (6 μg/mL) was measured. To test the effect of heparin on the interaction, C3d was coupled onto the wells, and 125I-labeled FH19–20 or FH19–20 mutant (6 μg/mL) was added in the presence of increasing amounts of dextran (Sigma-Aldrich) or heparin decasaccharide (Neoparin, Inc.); dextran had no effect (Fig. S6). The assays were performed in 1% BSA in PBS or 1/2 PBS (70 mM NaCl, 5 mM phosphate).
Cell Binding Analyses.
Binding of 125I-FH19–20 or FH19–20 mutants to C3b-coated or uncoated sheep erythrocytes was measured after coating the cells with approximately 25,000 C3b molecules/cell using purified C3, B, and D as previously described (29). The amount of cell-bound C3b was calculated using a trace amount of 125I-labeled C3 in the reaction mixtures. The C3b-coated cells were incubated with 125I-FH19–20 or FH19–20 mutants (2 μg/mL) for 2 h at 37 °C. The assay was performed in 1% BSA in veronal-buffered saline (VBS) as previously described (14).
Statistical Analyses.
Statistics and error bars are shown for independent experiments. Data were subjected to one-way ANOVA (n = 3 or 4; α-levels 0.05, 0.01, and 0.001) followed by Dunnett's posttest to compare the mean values obtained with the WT protein vs. the mutant proteins using GraphPad Prism 5.01 software.
Acknowledgments
We thank Seija Mäki and Katja Rosti for technical assistance in crystal screening and Marjatta Ahonen and Kirsti Widing for technical assistance in protein expression. T.K. is funded by the Academy of Finland. M.J.L. was partially funded by the Maud Kuistila Memorial Foundation, the Emil Aaltonen Foundation, and the Finnish Cultural Foundation. S.M. and T.S.J. are funded by the Academy of Finland, the Sigrid Jusélius Foundation, and the Helsinki University Hospital Funds (EVO). A.G. is funded by the Academy of Finland and the Sigrid Jusélius Foundation.
Footnotes
The authors declare no conflict of interest.
Data deposition: The crystallography, atomic coordinates, and structure factors have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2XQW).
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1017087108/-/DCSupplemental.
References
- 1.Meri S, Pangburn MK. Discrimination between activators and nonactivators of the alternative pathway of complement: Regulation via a sialic acid/polyanion binding site on factor H. Proc Natl Acad Sci USA. 1990;87:3982–3986. doi: 10.1073/pnas.87.10.3982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fearon DT. Regulation by membrane sialic acid of β1H-dependent decay-dissociation of amplification C3 convertase of the alternative complement pathway. Proc Natl Acad Sci USA. 1978;75:1971–1975. doi: 10.1073/pnas.75.4.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pangburn MK, Müller-Eberhard HJ. Complement C3 convertase: Cell surface restriction of β1H control and generation of restriction on neuraminidase-treated cells. Proc Natl Acad Sci USA. 1978;75:2416–2420. doi: 10.1073/pnas.75.5.2416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Manuelian T, et al. Mutations in factor H reduce binding affinity to C3b and heparin and surface attachment to endothelial cells in hemolytic uremic syndrome. J Clin Invest. 2003;111:1181–1190. doi: 10.1172/JCI16651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gordon DL, Kaufman RM, Blackmore TK, Kwong J, Lublin DM. Identification of complement regulatory domains in human factor H. J Immunol. 1995;155:348–356. [PubMed] [Google Scholar]
- 6.Kühn S, Skerka C, Zipfel PF. Mapping of the complement regulatory domains in the human factor H-like protein 1 and in factor H1. J Immunol. 1995;155:5663–5670. [PubMed] [Google Scholar]
- 7.Pangburn MK. Cutting edge: Localization of the host recognition functions of complement factor H at the carboxyl-terminal: Implications for hemolytic uremic syndrome. J Immunol. 2002;169:4702–4706. doi: 10.4049/jimmunol.169.9.4702. [DOI] [PubMed] [Google Scholar]
- 8.Jokiranta TS, et al. Binding of complement factor H to endothelial cells is mediated by the carboxy-terminal glycosaminoglycan binding site. Am J Pathol. 2005;167:1173–1181. doi: 10.1016/S0002-9440(10)61205-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jokiranta TS, Hellwage J, Koistinen V, Zipfel PF, Meri S. Each of the three binding sites on complement factor H interacts with a distinct site on C3b. J Biol Chem. 2000;275:27657–27662. doi: 10.1074/jbc.M002903200. [DOI] [PubMed] [Google Scholar]
- 10.Blackmore TK, et al. Identification of the second heparin-binding domain in human complement factor H. J Immunol. 1998;160:3342–3348. [PubMed] [Google Scholar]
- 11.Ram S, et al. A novel sialic acid binding site on factor H mediates serum resistance of sialylated Neisseria gonorrhoeae. J Exp Med. 1998;187:743–752. doi: 10.1084/jem.187.5.743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kavanagh D, Goodship TH. Atypical hemolytic uremic syndrome. Curr Opin Hematol. 2010;17:432–438. doi: 10.1097/MOH.0b013e32833cae86. [DOI] [PubMed] [Google Scholar]
- 13.Jokiranta TS, et al. Structure of complement factor H carboxyl-terminus reveals molecular basis of atypical haemolytic uremic syndrome. EMBO J. 2006;25:1784–1794. doi: 10.1038/sj.emboj.7601052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lehtinen MJ, Rops AL, Isenman DE, van der Vlag J, Jokiranta TS. Mutations of factor H impair regulation of surface-bound C3b by three mechanisms in atypical hemolytic uremic syndrome. J Biol Chem. 2009;284:15650–15658. doi: 10.1074/jbc.M900814200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ferreira VP, et al. The binding of factor H to a complex of physiological polyanions and C3b on cells is impaired in atypical hemolytic uremic syndrome. J Immunol. 2009;182:7009–7018. doi: 10.4049/jimmunol.0804031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Nagar B, Jones RG, Diefenbach RJ, Isenman DE, Rini JM. X-ray crystal structure of C3d: A C3 fragment and ligand for complement receptor 2. Science. 1998;280:1277–1281. doi: 10.1126/science.280.5367.1277. [DOI] [PubMed] [Google Scholar]
- 17.Lawrence MC, Colman PM. Shape complementarity at protein/protein interfaces. J Mol Biol. 1993;234:946–950. doi: 10.1006/jmbi.1993.1648. [DOI] [PubMed] [Google Scholar]
- 18.Frémeaux-Bacchi V, et al. Mutations in complement C3 predispose to development of atypical hemolytic uremic syndrome. Blood. 2008;112:4948–4952. doi: 10.1182/blood-2008-01-133702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maga TK, Nishimura CJ, Weaver AE, Frees KL, Smith RJ. Mutations in alternative pathway complement proteins in American patients with atypical hemolytic uremic syndrome. Hum Mutat. 2010;31:E1445–E1460. doi: 10.1002/humu.21256. [DOI] [PubMed] [Google Scholar]
- 20.Bhattacharjee A, Lehtinen MJ, Kajander T, Goldman A, Jokiranta TS. Both domain 19 and domain 20 of factor H are involved in binding to complement C3b and C3d. Mol Immunol. 2010;47:1686–1691. doi: 10.1016/j.molimm.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 21.Janssen BJ, Christodoulidou A, McCarthy A, Lambris JD, Gros P. Structure of C3b reveals conformational changes that underlie complement activity. Nature. 2006;444:213–216. doi: 10.1038/nature05172. [DOI] [PubMed] [Google Scholar]
- 22.Wu J, et al. Structure of complement fragment C3b-factor H and implications for host protection by complement regulators. Nat Immunol. 2009;10:728–733. doi: 10.1038/ni.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbert AP, Uhrín D, Lyon M, Pangburn MK, Barlow PN. Disease-associated sequence variations congregate in a polyanion recognition patch on human factor H revealed in three-dimensional structure. J Biol Chem. 2006;281:16512–16520. doi: 10.1074/jbc.M513611200. [DOI] [PubMed] [Google Scholar]
- 24.Hellwage J, et al. Complement C3b/C3d and cell surface polyanions are recognized by overlapping binding sites on the most carboxyl-terminal domain of complement factor H. J Immunol. 2002;169:6935–6944. doi: 10.4049/jimmunol.169.12.6935. [DOI] [PubMed] [Google Scholar]
- 25.Nilsson B, Nilsson Ekdahl K, Avila D, Nilsson UR, Lambris JD. Neoantigens in complement component C3 as detected by monoclonal antibodies. Mapping of the recognized epitopes by synthetic peptides. Biochem J. 1990;268:55–61. doi: 10.1042/bj2680055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nishida N, Walz T, Springer TA. Structural transitions of complement component C3 and its activation products. Proc Natl Acad Sci USA. 2006;103:19737–19742. doi: 10.1073/pnas.0609791104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cheng ZZ, et al. Complement factor H as a marker for detection of bladder cancer. Clin Chem. 2005;51:856–863. doi: 10.1373/clinchem.2004.042192. [DOI] [PubMed] [Google Scholar]
- 28.Isenman DE, Leung E, Mackay JD, Bagby S, van den Elsen JM. Mutational analyses reveal that the staphylococcal immune evasion molecule Sbi and complement receptor 2 (CR2) share overlapping contact residues on C3d: Implications for the controversy regarding the CR2/C3d cocrystal structure. J Immunol. 2010;184:1946–1955. doi: 10.4049/jimmunol.0902919. [DOI] [PubMed] [Google Scholar]
- 29.Koistinen V. Effect of complement-protein-C3b density on the binding of complement factor H to surface-bound C3b. Biochem J. 1991;280:255–259. doi: 10.1042/bj2800255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 32.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method. Acta Crystallogr D Biol Crystallogr. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 33.Adams PD, et al. PHENIX: A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]