Abstract
All animals detect osmotic and mechanical stimuli, but the molecular basis for these responses is incompletely understood. The vertebrate transient receptor potential channel vanilloid subfamily 4 (TRPV4) (VR-OAC) cation channel has been suggested to be an osmo/mechanosensory channel. To assess its function in vivo, we expressed TRPV4 in Caenorhabditis elegans sensory neurons and examined its ability to generate behavioral responses to sensory stimuli. C. elegans ASH neurons function as polymodal sensory neurons that generate a characteristic escape behavior in response to mechanical, osmotic, or olfactory stimuli. These behaviors require the TRPV channel OSM-9 because osm-9 mutants do not avoid nose touch, high osmolarity, or noxious odors. Expression of mammalian TRPV4 in ASH neurons of osm-9 worms restored avoidance responses to hypertonicity and nose touch, but not the response to odorant repellents. Mutations known to reduce TRPV4 channel activity also reduced its ability to direct nematode avoidance behavior. TRPV4 function in ASH required the endogenous C. elegans osmotic and nose touch avoidance genes ocr-2, odr-3, osm-10, and glr-1, indicating that TRPV4 is integrated into the normal ASH sensory apparatus. The osmotic and mechanical avoidance responses of TRPV4-expressing animals were different in their sensitivity and temperature dependence from the responses of wild-type animals, suggesting that the TRPV4 channel confers its characteristic properties on the transgenic animals' behavior. These results provide evidence that TRPV4 can function as a component of an osmotic/mechanical sensor in vivo.
Animals perceive and avoid danger through sensory nociception, the detection of noxious stimuli in the external environment or noxious conditions in internal tissues. Noxious stimuli are polymodal: a stimulus can be recognized as noxious based on its specific chemical composition, as in inflammatory cytokines, based on its physicochemical properties such as pH and osmolarity, based on purely physical properties like pressure or heat, or based on a combination of properties. Some types of nociceptive neurons are intrinsically polymodal, detecting many noxious stimuli, whereas others are specialized to detect particular physical or chemical stimuli (1, 2). In mammals, dorsal root ganglion neurons sense noxious chemical, thermal, osmotic, acidic, or physical cues, or a combination of such cues (3). The more specific inner ear hair cells sense sound and acceleration (4), and neurons in circumventricular organs of the brain sense systemic osmotic pressure (5, 6). Although sensory transduction mechanisms for chemosensation and thermosensation have been extensively characterized, the molecular mechanisms for sensing mechanical or physicochemical stimuli are less well understood.
Nociception has been genetically characterized in the invertebrates Drosophila melanogaster and Caenorhabditis elegans. In these organisms, mutants with deficits in the response to noxious mechanical, osmotic, and chemical stimuli have been characterized (1, 7). In C. elegans, genetic studies have identified two distinct mechanosensory nociceptive pathways (8, 9). In one mechanosensory pathway, a mechanical stimulus applied to the animal's body elicits a withdrawal response. Members of the mechanically activated channel, degenerin channel, epithelial sodium channel (MEC/DEG/EnaC) family are required for body touch mechanosensation, and are likely to form the mechanosensory channels in these neurons (8, 10, 11). In another mechanosensory pathway, an avoidance response to nose touch is mediated by the ciliated ASH sensory neurons in the head (9, 12). ASH is a polymodal nociceptor: stimulation of the ASH cilia by either light touch, a hyperosmolar solution, or repulsive odorants leads the animal to reverse its direction of movement (13). Animals with mutations in osm-9 or ocr-2 do not avoid any of these stimuli. The osm-9 and ocr-2 genes encode putative ion channels belonging to the transient receptor potential (TRP) channel superfamily, vanilloid subfamily (TRPV) (14–16) and are proposed to encode the sensory transduction channel in ASH (17). The extent to which OSM-9 responds directly to sensory stimuli or indirectly to signal transduction pathways is unknown. Interestingly, the Drosophila ortholog of the ocr genes is required for fly hearing, a mechanosensory process (18). Another TRP family channel, the Drosophila nompC channel, is also thought to encode a mechanosensory channel, and a zebrafish nompC channel is implicated in mechanosensory hair cell function (19, 20).
TRPV4, also known as vanilloid receptor related osmotically activated channel (VR-OAC), OTRPC4, TRP12, or VRL-2, is a vertebrate nonspecific cation channel that is gated by osmotic stimuli in heterologous expression systems (15, 21–25). TRPV4 belongs to the TRPV subfamily of the TRP ion channel superfamily (15, 16, 25, 26) and shows similarity to the C. elegans channels OSM-9 (26% amino acid identity, 44% identity or conservative change) and OCR-2 (24% identity, 38% identity or conservative change). TRPV4 is also related to the vertebrate vanilloid receptor 1 (VR1; TRPV1), the vanilloid receptor-like channel (VRL-1; TRPV2), and the recently reported TRPV3 (27–32). TRPV1–3 are cation channels that are gated by warm and hot thermal stimuli.
The expression of TRPV4 in the circumventricular organs of the vertebrate CNS suggests that this channel could sense systemic osmotic pressure. In transfected cells, TRPV4 is gated by hypotonicity within the physiological range (22, 23), and hypotonicity-gated TRPV4 function is suggested as one mechanism of pain sensation (33). However, in the circumventricular organ, hypertonicity is thought to be the physiological stimulus. This difference could result from accessory molecules in osmosensitive cells that modify TRPV4 function. TRPV4 is also expressed in vertebrate mechanosensory cells (1). Most models of mechanosensation propose the existence of multiple accessory proteins to mechanosensory channels (6). A full understanding of TRPV4 function might best be accomplished in cells with intrinsic mechanosensory or osmosensory function, rather than entirely heterologous cells.
To better understand the function of TRPV4 in vivo, we have expressed rat TRPV4 in the ASH neurons of C. elegans. Our results indicate that the rat channel can direct osmotic and mechanosensory behaviors when integrated into the normal ASH sensory transduction apparatus.
Methods
C. elegans Strains and Transgenic Animals. C. elegans was grown at 20°C and maintained by using standard methods (34, 35). Wild-type animals were C. elegans variety Bristol, strain N2. Mutations used in this work were osm-10(n1602) III, glr-1(ky176) III, osm-9(ky10) IV, ocr-2(ak47) IV, and odr-3(n1605) V. All are strong loss-of-function mutations that are believed to represent null alleles. Transgenic arrays used were kyEx575(sra-6::trpv4 elt-2::GFP), kyEx594(sra-6::trpv4::GFP odr-1::dsRED), kyEx609(sra-6::trpv4ΔN elt-2::GFP), kyEx596(sra6::trpv4ΔC elt-2::GFP), kyEx608(sra6::trpv4ΔNΔC elt-2::GFP), kyEx612(sra-6::trpv4 D672K elt-2::GFP), kyEx597 and kyEx605(sra-6::trpv4 M680K elt-2::GFP), kyEx598 and kyEx611(sra-6::trpv4 D682K elt-2::GFP).
The coding region of the rat TRPV4 cDNA was expressed under the sra-6 promoter in the C. elegans expression vector pPD49.26 (17). This promoter directs strong expression in the ASH and PVQ neurons, and weak expression in ASI neurons (35). Germ-line transformation was carried out by injecting DNA at 50 ng/μl (35) together with the intestinal elt-2::GFP marker at 10 ng/μl (17). Transgenic lines were maintained in an osm-9(ky10) genetic background; ky10 is a null allele of osm-9 that results from an early stop codon.
Details of molecular biology are available upon request. OSM-9::GFP5 lines were as described (34).
Neurons were identified for laser ablation by using differential interference contrast optics and a combination of positional and morphological cues as described (36). Cell kills of ASH were confirmed by bilateral absence of dye-filling with 1,1′-dioctadecyl-3,3,3′,3′-tetramethylindodicarbocyanine perchlorate (DiD).
Imaging Animals expressing sra-6::trpv4::gfp, a C-terminally tagged protein, were analyzed with the Deltavision imaging system, which deconvolutes multiple sections of fluorescent micrographs to generate a projection and a 3D reconstruction (37).
Immunohistochemistry of fixed animals was performed by using an antibody raised against the first 445 aa of TRPV4 (a generous gift from Stefan Heller, Harvard University, Boston) and secondary goat anti-rabbit FITC antisera by using described methods (38).
Behavioral Assays. All behavioral assays were performed by investigators blinded to the genotype of the animals. Osmotic avoidance assays were performed by exposing individual animals to a drop of 1 M fructose or glycerol (39). A graded series of osmotic stimuli was obtained through serial dilution of 1 M glycerol. A reversal of more than half a body length before the animal had left the drop area was scored as a response (17). For nose touch response, the tip of the nose of a forward-moving animal was touched with a fine hair, and reversal was scored as a response (9, 13). For chemical avoidance, 2-octanone and 1-octanol were used. A microcapillary containing 5 μl of 2-octanone or 1-octanol was placed immediately in front of a freely moving adult animal (17), and reversal within 3 s was scored as a response. For all avoidance assays, 10 trials per animal were recorded and the percentage of positive responses for each animal was compiled per group.
Temperature was modulated by using an over-the-counter heat lamp (75-W bulb) at a distance of 10–12 cm from the center of the assay plate under the dissecting microscope. Plate temperature was recorded and calibrated with a precision thermometer (YSI Temperature, Dayton, OH).
For further details, see Supporting Methods, which is published as supporting information on the PNAS web site, www.pnas.org.
Tissue Culture. Cultivation of Chinese hamster ovary (CHO) cells, transfection, and stimulation were carried out as described (22). For further details, see Supporting Methods.
Statistical Analysis. Pairwise comparison was performed by t test, multigroup comparison by ANOVA analysis in combination with Dunnett's posttest analysis. The statistical program PRISM4 for Macintosh was used (GraphPad, San Diego).
Results and Discussion
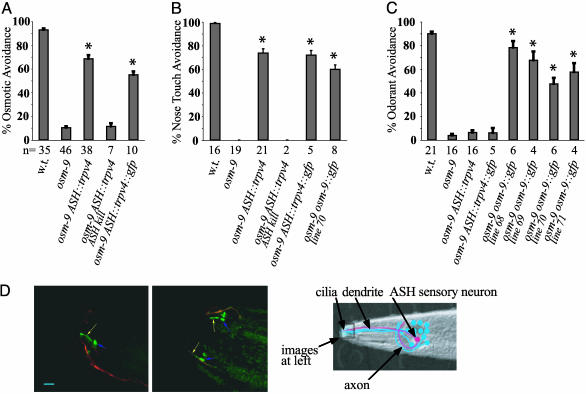
To establish the properties of TRPV4 in a sensory system in vivo, we tested its ability to function within the ciliated ASH sensory neurons of C. elegans in the presence of an osm-9 mutation that eliminates all endogenous ASH functions. osm-9 animals are almost completely defective in their response to noxious hyperosmotic stimuli, nose touch and several aversive olfactory stimuli. By contrast, transgenic animals expressing TRPV4 or TRPV4::GFP in ASH neurons avoided both osmotic stimuli (P < 0.01) and nose touch (P < 0.01) (Fig. 1 A and B and Movies 1–3, which are published as supporting information on the PNAS web site). The osmotic avoidance response of osm-9 ASH::trpv4 animals was slightly delayed relative to the endogenous C. elegans behavior; this result is consistent with the relatively slow activation of vertebrate osmosensory channels.
Fig. 1.
TRPV4 expression directs osmotic and nose touch avoidance in osm-9 mutants. (A) Osmotic avoidance of 1 M fructose or glycerol. (B) Nose touch avoidance. (C) Avoidance of the odorant 2-octanone. Wild-type (w.t.) and osm-9(ky10) animals with or without an ASH::trpv4 transgene, an ASH::trpv4::gfp transgene, or an osm-9::gfp5 transgene were tested. “ASH kill” denotes bilateral laser ablation of the ASH neuron. In all panels, asterisks denote significant differences between the indicated group and the osm-9 group (P < 0.01, one-way ANOVA with Dunnett's posttest analysis). Error bars denote SEM. n = number of animals tested, 10 trials each. (D) TRPV4::GFP expression in the nociceptive ASH neurons. (Left) Lateral view. (Center) Dorsal view. TRPV4::GFP in ASH appears green, and an ODR-1::dsRED fusion protein expressed in the dendrite and weakly in the cilium of the adjacent AWC sensory neuron appears red. Yellow arrow, ASH cilia; blue arrow, base of dendrite. (Scale bar = 5 μm.) (Right) Schematic diagram of C. elegans ASH sensory neuron (red), 1 of 12 amphid sensory neurons (blue) that extend dendrites to the nose, where they terminate in sensory cilia. Two amphids each contain an ASH polymodal nociceptive neuron; only the left amphid is shown. The area depicted in the fluorescent micrographs is highlighted.
To confirm that osmotic avoidance and nose touch avoidance were generated by TRPV4 expression in ASH, the ASH neurons were ablated in osm-9 ASH::trpv4 animals by using a laser microbeam. Ablation of the ASH neurons eliminated osmotic avoidance and nose touch avoidance in the transgenic strain (Fig. 1 A and B).
The biologically active TRPV4::GFP protein was localized to the cilia of ASH, where mechanical and osmotic stimuli are sensed (Fig. 1D). It was also present in ASH cell bodies, but not in axons or dendrites. Both OSM-9 and OCR-2 are similarly localized to sensory cilia (17).
TRPV4 failed to restore ASH-mediated odorant avoidance of the volatile repellents 2-octanone and 1-octanol to osm-9 mutants (Fig. 1C and data not shown). Thus, the TRPV4 channel appears to confer osmosensitivity and mechanosensitivity, but not chemosensitivity, to the ASH neuron in osm-9 mutants. This property is consistent with the expected functions of TRPV4, and different from the normal functions of OSM-9. As a control for this experiment, we tested an osm-9 strain rescued with osm-9::gfp in parallel to the osm-9 ASH::trpv4 and osm-9 ASH::trpv4::gfp strains. osm-9 [osm-9::gfp]+ animals were rescued for osmosensation, mechanosensation, and odor sensation, unlike osm-9 ASH::trpv4 animals. The difference between OSM-9 and TRPV4 in odorant response appears to be qualitative rather than quantitative. The osm-9 [osm-9::gfp]+ strain exhibited relatively weak rescue of nose touch compared with the ASH::trpv4 line (Fig. 1B), presumably because of lower expression of the transgene, yet the line still rescued odorant avoidance (Fig. 1C). Thus, the specific rescue of the osmotic and mechanical sensing deficits of osm-9 by TRPV4 represents an intrinsic difference between TRPV4 and OSM-9.
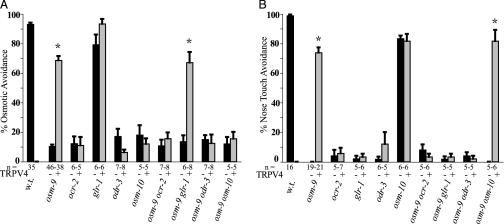
TRPV4-expressing animals avoided hyperosmotic stimuli, but in heterologous expression systems, TRPV4-expressing cells are activated only in response to hypoosmolar stimuli (22, 23). This difference suggests that the ASH sensory neuron might contain accessory molecules that allow TRPV4 to sense hyperosmotic stimuli. ocr-2, odr-3, and osm-10 genes are required for endogenous ASH osmosensory behaviors. ocr-2 encodes another TRPV ion channel subunit, odr-3 encodes a Gα-protein (40), and osm-10 encodes a cytoplasmic protein in the ASH neuron (41). To assess whether these genes were also required for TRPV4 function in ASH neurons, osm-9 ASH::trpv4 transgenic animals were crossed to animals carrying ocr-2, odr-3, and osm-10 mutations. TRPV4 was unable to generate osmosensory behaviors in ocr-2, odr-3, and osm-10 mutants (Fig. 2A). Thus, endogenous osmosensory molecules in ASH collaborate with TRPV4 to generate an osmotic response. OCR-2 is required for OSM-9 localization to ASH sensory cilia, but is not required for cilia localization of TRPV4 (Fig. 5, which is published as supporting information on the PNAS web site). This result suggests that the requirement for both OCR-2 and OSM-9 or TRPV4 reflects activity of both channel subunits, and not only their mutual localization to cilia.
Fig. 2.
TRPV4 functions with endogenous C. elegans osmo- and mechanosensory genes. (A) Osmotic avoidance of 1 M fructose or glycerol by single mutants (Left) or double mutants with osm-9 (Right), with (gray) or without (black) the ASH::trpv4 transgene. The first three bars represent the same results depicted in Fig. 1 A. (B) Nose touch avoidance. Strains as are in A. The first three bars represent the same results depicted in Fig. 1B. In both panels, asterisks denote statistically significant differences between the trpv4 transgenic group and the parallel nontransgenic group (P < 0.01, one-way ANOVA with Dunnett's posttest analysis). n = number of animals tested, 10 trials each; w.t., wild type.
Endogenous ASH mechanosensation requires ocr-2 and odr-3, but not osm-10. ASH mechanosensation also requires the glr-1 AMPA-type glutamate receptor, which is not required for osmosensation (42, 43). Mechanosensation mediated by ASH::trpv4 required the function of ocr-2, odr-3, and glr-1 genes (Fig. 2B). As in the endogenous response, glr-1 was not required for ASH::trpv4-mediated osmosensation, and osm-10 was not required for ASH::trpv4-mediated mechanosensation.
Expression of TRPV1 (VR1) does not rescue osmosensation or mechanosensation in osm-9 mutants, but does confer a strong avoidance behavior to capsaicin, a TRPV1 ligand (17). The capsaicin response of osm-9 ASH::trpv1 transgenic animals was retained in ocr-2, odr-3, osm-10, and glr-1 mutant backgrounds. Thus, TRPV4, but not TRPV1, exploits all of the characterized endogenous osmosensory and mechanosensory signaling proteins in ASH to generate behavior.
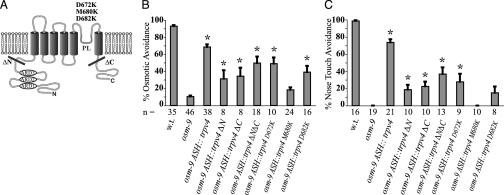
If TRPV4 acts as a mechanosensory or osmosensory channel, its behavioral functions should correlate with its ability to function as an ion channel. Three point mutations in the predicted pore–loop domain of TRPV4, D672K, M680K, and D682K were made based on published studies of other family members, TRPV1 (VR1) and TRPV5 (EcaC) (Fig. 3A and Fig. 6, which is published as supporting information on the PNAS web site) (44, 45). The replacement of a methionine with lysine at position 680 (M680K) eliminated the ability of TRPV4 to confer nose touch avoidance and osmotic avoidance to the osm-9 mutant (Fig. 3 B and C). TRPV4M680K lacked channel activity when expressed in mammalian tissue culture cells stimulated with a TRPV4-activating phorbol ester or hypotonic solution (Fig. 7, which is published as supporting information on the PNAS web site). This observation is consistent with previous studies showing that residue M680 is required for TRPV ion channel function (44–46). TRPV4-mediated behaviors were greatly diminished in animals expressing TRPV4 mutations at position 682 (D682K) and at position 672 (D672K) (Fig. 3 B and C). Deletion of the TRPV4 N terminus (residues 1–410), C terminus (residues 741–871), or both N- and C-termini reduced but did not eliminate its behavioral function (Fig. 3 B and C). The ΔNΔC mutant lacking both N and C termini localized to the ASH cilia, as did the M680K mutant channel (Fig. 8, which is published as supporting information on the PNAS web site).
Fig. 3.
TRPV4 mutations affect C. elegans behavior and channel function. (A) Schematic drawing of TRPV4 including sites of mutations. ARD, ankyrin-repeat domain; PL, pore-loop domain; ΔN, extent of deleted N-terminal domain; ΔC, extent of deleted C-terminal domain. N and C termini are intracellular. (B) Osmotic avoidance of 1 M fructose or glycerol. Bar graphs depict pooled data of three independent transgenic lines for each mutant. The first three bars represent the same results depicted in Fig. 1 A. (C) Nose touch avoidance. Bar graphs depict the combined results for three independent transgenic lines of each mutant. The first three bars represent the same results depicted in Fig. 1B. For B and C, asterisk denotes significant differences between the indicated group and the osm-9 group (one-way ANOVA with Dunnett's posttest analysis; for B: P < 0.01 except ΔN group and ΔC group, here P < 0.05; for C: P < 0.01 except ΔN group). n = number of animals tested, 10 trials each; w.t., wild type.
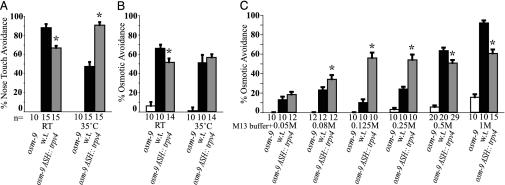
TRPV4 osmotic responses are enhanced at physiological temperatures in transfected mammalian cells (22, 47). We assessed the temperature-dependence of avoidance behaviors of wild-type and osm-9 ASH::trpv4 animals at room temperature (23°C) and 34–35°C, the upper limit of C. elegans viability. Wild-type animals responded significantly more frequently to nose touch at room temperature, whereas osm-9 ASH::trpv4 responded more strongly at 35°C (Fig. 4A and Fig. 9, which is published as supporting information on the PNAS web site); a smaller effect was observed for osmotic avoidance (Fig. 4B). Although TRPV4-induced behaviors appeared to reflect the temperature modulation of TRPV4, TRPV4 did not generate avoidance behavior to thermal stimuli alone (48, 49), whereas TRPV1, a channel implicated in noxious heat responses, conferred a significant thermal avoidance response to isotonic M13 buffer at 37°C (Fig. 10, which is published as supporting information on the PNAS web site). Thus, the transduction of osmotic and mechanical stimuli in osm-9 ASH::trpv4 animals bears the molecular signature of TRPV4. Somatosensory perception of mechanical stimuli in humans and seals is modulated by temperature (50–52), and TRPV4 may recapitulate this effect at the molecular level.
Fig. 4.
TRPV4 avoidance behaviors are modulated by stimulus strength and temperature. (A) Nose touch avoidance at different temperatures. (B) Osmotic avoidance of 0.5 M glycerol at different temperatures. (C) Osmotic avoidance of different concentrations of glycerol. The osmotic strength of M13 buffer is 295 mOsmol/liter; i.e., a 0.05 M osmotic stimulus yields a final concentration of 345 mOsmol/liter. Asterisks denote significant differences between osm-9 ASH::trpv4 and wild-type (w.t.) groups (P < 0.05, t test for the 0.08 M group in C, P < 0.01 for all other groups in A–C). n = number of animals tested, 10 trials each.
Vertebrate osmosensation in the central nervous system is exquisitely sensitive to small increases in systemic osmolarity. By contrast, C. elegans osmosensation detects relatively large increases in external osmolarity that may be associated with salty or brackish water. This difference in physiological function might predict a difference in the threshold of osmosensory channels in C. elegans and vertebrates. The osmotic avoidance behaviors of wild-type, osm-9, and osm-9 ASH::trpv4 animals were assessed by using solutions with different osmotic strengths (Fig. 4C). Wild-type C. elegans responded weakly to increased osmolalities of 0.25 M and below, with a maximal behavioral response at a 1 M osmotic stimulus. By contrast, osm-9 ASH::trpv4 animals responded equally well to 0.125 M, 0.25 M, 0.5 M, and 1 M osmotic stimuli. Thus, the osm-9 ASH::trpv4 animals exhibited a significantly lower threshold to hyperosmotic stimuli than wild-type animals, as would be consistent with the high sensitivity of vertebrate osmosensation.
TRPV4 allows ASH neurons to respond to hypertonicity, but it responds to hypotonicity when expressed in transfected cells. One possible explanation for this difference is suggested by the properties of the mechanosensitive ion channel gramicidin A, which behaves either as a stretch-inactivated or as a stretch-activated channel depending on the lipid composition of the surrounding lipid bilayer (53). An alternate possibility is that TRPV4 forms heteromultimeric complexes with C. elegans proteins. TRP ion channels can form heteromeric complexes with related family members (17, 25, 54, 55); notably, the two TRPV channels OCR-2 and OSM-9 may associate in ASH (17).
Expression of rat TRPV4 in ASH rescued the transduction of osmotic and mechanical stimuli in osm-9 but not ocr-2 mutants, whereas rat TRPV1 did not rescue either mutant. These results suggest that TRPV4 and OSM-9 have orthologous functions, indicating a phylogenetic conservation of function. It is interesting that TRPV4 could not rescue the odorant avoidance deficit of osm-9 mutants for the two odors tested. C. elegans senses odorants by using G protein-coupled receptors; it is possible that TRPV4 does not interact with an essential component of the G protein-coupled receptor signaling pathway, although it does interact with the components required for sensing physical stimuli. Among several models consistent with these results (Fig. 11, which is published as supporting information on the PNAS web site), the conserved role of TRPV ion channels in osmosensory pathways suggests a central, perhaps direct, role for these channels in osmosensation and mechanosensation. However, the essential role of the G protein α subunit ODR-3 in mechanosensation and chemosensation leaves open the possible involvement of G protein-coupled receptors as mediators or regulators of these sensory signals.
Our results provide evidence that mammalian TRPV4 can function as a central component of the sensor for osmotic and mechanical stimuli in ASH sensory neurons in C. elegans. Expression of TRPV4 directs a behavioral response to osmotic and mechanical stimuli in vivo, and specific properties of the behavior are conferred by the mammalian channel. This conclusion is consistent with findings in trpv4 null mice (56–58), and supports the hypothesis that TRPV4 functions as part of a mammalian osmotic and mechanical sensor.
Supplementary Material
Acknowledgments
The Rockefeller University Bio-Imaging Resource Center (Alison North, director), provided assistance with imaging, and Jim Hudspeth, Shai Shaham (both from The Rockefeller University), Justin Blau (New York University, New York), Stefan Heller (Harvard University, Boston) and Sebastian Martinek (Pennie Partners, New York) provided valuable suggestions and support. W.L., C.I.B., and J.M.F. were supported by the National Institutes of Health. C.I.B. and J.M.F. are Investigators with the Howard Hughes Medical Institute. D.M.T. received a National Science Foundation predoctoral fellowship.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Chemical Communication in a Post-Genomic World,” held January 17–19, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: TRPV, transient receptor potential channel, vanilloid subfamily; VRL-1, vanilloid receptor-like receptor 1; VR1, vanilloid receptor 1.
References
- 1.Goodman, M. B. & Schwarz, E. M. (2003) Annu. Rev. Physiol. 65, 429-452. [DOI] [PubMed] [Google Scholar]
- 2.Gillespie, P. G. & Walker, R. G. (2001) Nature 413, 194-202. [DOI] [PubMed] [Google Scholar]
- 3.Gardner, E. P., Martin, J. H. & Jessell, T. M. (2000) in Principles of Neural Science, ed. Kandel E. R., Schwartz, J. H. & Jessell, T. M. (McGraw–Hill, New York), pp. 430-450.
- 4.Hudspeth, A. J. (1989) Nature 341, 397-404. [DOI] [PubMed] [Google Scholar]
- 5.Bourque, C. W. & Oliet, S. H. (1997) Annu. Rev. Physiol. 59, 601-619. [DOI] [PubMed] [Google Scholar]
- 6.Denton, D. A., McKinley, M. J. & Weisinger, R. S. (1996) Proc. Natl. Acad. Sci. USA 93, 7397-7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Duggan, A., Garcia-Anoveros, J. & Corey, D. P. (2000) Curr. Biol. 10, R384-R387. [DOI] [PubMed] [Google Scholar]
- 8.Goodman, M. B., Ernstrom, G. G., Chelur, D. S., O'Hagan, R., Yao, C. A. & Chalfie, M. (2002) Nature 415, 1039-1042. [DOI] [PubMed] [Google Scholar]
- 9.Kaplan, J. M. & Horvitz, H. R. (1993) Proc. Natl. Acad. Sci. USA 90, 2227-2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Garcia-Anoveros, J. & Corey, D. P. (1997) Annu. Rev. Neurosci. 20, 567-594. [DOI] [PubMed] [Google Scholar]
- 11.Price, M. P., Lewin, G. R., McIlwrath, S. L., Cheng, C., Xie, J., Heppenstall, P. A., Stucky, C. L., Mannsfeldt, A. G., Brennan, T. J., Drummond, H. A., et al. (2000) Nature 407, 1007-1011. [DOI] [PubMed] [Google Scholar]
- 12.Bargmann, C. I. & Kaplan, J. M. (1998) Annu. Rev. Neurosci. 21, 279-308. [DOI] [PubMed] [Google Scholar]
- 13.Bargmann, C. I., Thomas, J. H. & Horvitz, H. R. (1990) Cold Spring Harbor Symp. Quant. Biol. 55, 529-538. [DOI] [PubMed] [Google Scholar]
- 14.Montell, C., Birnbaumer, L., Flockerzi, V., Bindels, R. J., Bruford, E. A., Caterina, M. J., Clapham, D. E., Harteneck, C., Heller, S., Julius, D., et al. (2002) Mol. Cell 9, 229-231. [DOI] [PubMed] [Google Scholar]
- 15.Clapham, D. E., Runnels, L. W. & Strubing, C. (2001) Nat. Rev. Neurosci. 2, 387-396. [DOI] [PubMed] [Google Scholar]
- 16.Gunthorpe, M. J., Benham, C. D., Randall, A. & Davis, J. B. (2002) Trends Pharmacol. Sci. 23, 183-191. [DOI] [PubMed] [Google Scholar]
- 17.Tobin, D., Madsen, D. M., Kahn-Kirby, A., Peckol, E., Moulder, G., Barstead, R., Maricq, A. V. & Bargmann, C. I. (2002) Neuron 35, 307-318. [DOI] [PubMed] [Google Scholar]
- 18.Kim, J., Chung, Y. D., Park, D. Y., Choi, S., Shin, D. W., Soh, H., Lee, H. W., Son, W., Yim, J., Park, C. S., et al. (2003) Nature 424, 81-84. [DOI] [PubMed] [Google Scholar]
- 19.Walker, R. G., Willingham, A. T. & Zuker, C. S. (2000) Science 287, 2229-2234. [DOI] [PubMed] [Google Scholar]
- 20.Sidi, S., Friedrich, R. W. & Nicolson, T. (2003) Science 301, 96-99. [DOI] [PubMed] [Google Scholar]
- 21.Delany, N. S., Hurle, M., Facer, P., Alnadaf, T., Plumpton, C., Kinghorn, I., See, C. G., Costigan, M., Anand, P., Woolf, C. J., et al. (2001) Physiol. Genomics 4, 165-174. [DOI] [PubMed] [Google Scholar]
- 22.Liedtke, W., Choe, Y., Marti-Renom, M. A., Bell, A. M., Denis, C. S., Sali, A., Hudspeth, A. J., Friedman, J. M. & Heller, S. (2000) Cell 103, 525-535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Strotmann, R., Harteneck, C., Nunnenmacher, K., Schultz, G. & Plant, T. D. (2000) Nat. Cell Biol. 2, 695-702. [DOI] [PubMed] [Google Scholar]
- 24.Wissenbach, U., Bodding, M., Freichel, M. & Flockerzi, V. (2000) FEBS Lett. 485, 127-134. [DOI] [PubMed] [Google Scholar]
- 25.Montell, C., Birnbaumer, L. & Flockerzi, V. (2002) Cell 108, 595-598. [DOI] [PubMed] [Google Scholar]
- 26.Harteneck, C., Plant, T. D. & Schultz, G. (2000) Trends Neurosci. 23, 159-166. [DOI] [PubMed] [Google Scholar]
- 27.Caterina, M. J., Schumacher, M. A., Tominaga, M., Rosen, T. A., Levine, J. D. & Julius, D. (1997) Nature 389, 816-824. [DOI] [PubMed] [Google Scholar]
- 28.Caterina, M. J., Rosen, T. A., Tominaga, M., Brake, A. J. & Julius, D. (1999) Nature 398, 436-441. [DOI] [PubMed] [Google Scholar]
- 29.Caterina, M. J., Leffler, A., Malmberg, A. B., Martin, W. J., Trafton, J., Petersen-Zeitz, K. R., Koltzenburg, M., Basbaum, A. I. & Julius, D. (2000) Science 288, 306-313. [DOI] [PubMed] [Google Scholar]
- 30.Peier, A. M., Reeve, A. J., Andersson, D. A., Moqrich, A., Earley, T. J., Hergarden, A. C., Story, G. M., Colley, S., Hogenesch, J. B., McIntyre, P., et al. (2002) Science 296, 2046-2049. [DOI] [PubMed] [Google Scholar]
- 31.Smith, G. D., Gunthorpe, M. J., Kelsell, R. E., Hayes, P. D., Reilly, P., Facer, P., Wright, J. E., Jerman, J. C., Walhin, J. P., Ooi, L., et al. (2002) Nature 418, 186-190. [DOI] [PubMed] [Google Scholar]
- 32.Xu, H., Ramsey, I. S., Kotecha, S. A., Moran, M. M., Chong, J. A., Lawson, D., Ge, P., Lilly, J., Silos-Santiago, I., Xie, Y., et al. (2002) Nature 418, 181-186. [DOI] [PubMed] [Google Scholar]
- 33.Allessandri-Haber, N., Yeh, J., Boyd, A. E., Parada, C. A., Chen, X., Reichling, D. B. & Levine, J. D. (2003) Neuron 39, 497-511. [DOI] [PubMed] [Google Scholar]
- 34.Colbert, H. A., Smith, T. L. & Bargmann, C. I. (1997) J. Neurosci. 17, 8259-8269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Troemel, E. R., Chou, J. H., Dwyer, N. D., Colbert, H. A. & Bargmann, C. I. (1995) Cell 83, 207-218. [DOI] [PubMed] [Google Scholar]
- 36.Bargmann, C. I. & Avery, L. (1995) Methods Cell Biol. 48, 225-250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chikashige, Y., Ding, D. Q., Funabiki, H., Haraguchi, T., Mashiko, S., Yanagida, M. & Hiraoka, Y. (1994) Science 264, 270-273. [DOI] [PubMed] [Google Scholar]
- 38.L'Etoile, N. D. & Bargmann, C. I. (2000) Neuron 25, 575-586. [DOI] [PubMed] [Google Scholar]
- 39.Hilliard, M. A., Bargmann, C. I. & Bazzicalupo, P. (2002) Curr. Biol. 12, 730-734. [DOI] [PubMed] [Google Scholar]
- 40.Roayaie, K., Crump, J. G., Sagasti, A. & Bargmann, C. I. (1998) Neuron 20, 55-67. [DOI] [PubMed] [Google Scholar]
- 41.Hart, A. C., Kass, J., Shapiro, J. E. & Kaplan, J. M. (1999) J. Neurosci. 19, 1952-1958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hart, A. C., Sims, S. & Kaplan, J. M. (1995) Nature 378, 82-85. [DOI] [PubMed] [Google Scholar]
- 43.Maricq, A. V., Peckol, E., Driscoll, M. & Bargmann, C. I. (1995) Nature 378, 78-81. [DOI] [PubMed] [Google Scholar]
- 44.Nilius, B., Vennekens, R., Prenen, J., Hoenderop, J. G., Droogmans, G. & Bindels, R. J. (2001) J. Biol. Chem. 276, 1020-1025. [DOI] [PubMed] [Google Scholar]
- 45.Garcia-Martinez, C., Morenilla-Palao, C., Planells-Cases, R., Merino, J. M. & Ferrer-Montiel, A. (2000) J. Biol. Chem. 275, 32552-32558. [DOI] [PubMed] [Google Scholar]
- 46.Voets, T., Prenen, J., Vriens, J., Watanabe, H., Janssens, A., Wissenbach, U., Boedding, M., Droogmans, G. & Nilius, B. (2002) J. Biol. Chem. 277, 33704-33710. [DOI] [PubMed] [Google Scholar]
- 47.Gao, X., Wu, L. & O'Neil, R. G. (2003) J. Biol. Chem. 278, 27129-27137. [DOI] [PubMed] [Google Scholar]
- 48.Guler, A. D., Lee, H., Iida, T., Shimizu, I., Tominaga, M. & Caterina, M. (2002) J. Neurosci. 22, 6408-6414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Watanabe, H., Vriens, J., Suh, S. H., Benham, C. D., Droogmans, G. & Nilius, B. (2002) J. Biol. Chem. 277, 47044-47051. [DOI] [PubMed] [Google Scholar]
- 50.Dehnhardt, G., Mauck, B. & Hyvarinen, H. (1998) J. Exp. Biol. 201, 3023-3029. [DOI] [PubMed] [Google Scholar]
- 51.Fucci, D., Crary, M. & Wilson, H. (1976) Percept. Mot. Skills 43, 263-266. [DOI] [PubMed] [Google Scholar]
- 52.Weitz, J. (1941) J. Exp. Psychol. 28, 21-36. [Google Scholar]
- 53.Martinac, B. & Hamill, O. P. (2002) Proc. Natl. Acad. Sci. USA 99, 4308-4312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Xu, X. Z., Chien, F., Butler, A., Salkoff, L. & Montell, C. (2000) Neuron 26, 647-657. [DOI] [PubMed] [Google Scholar]
- 55.Xu, X. Z., Li, H. S., Guggino, W. B. & Montell, C. (1997) Cell 89, 1155-1164. [DOI] [PubMed] [Google Scholar]
- 56.Suzuki, M., Mizuno, A., Kodaira, K. & Imai, M. (2003) J. Biol. Chem. 278, 22664-22668. [DOI] [PubMed] [Google Scholar]
- 57.Mizuno, A., Matsumoto, N., Imai, M. & Suzuki, M. (2003) Am. J. Physiol. 285, C96-C101. [DOI] [PubMed] [Google Scholar]
- 58.Liedtke, W. & Friedman, J. M. (2003) Proc. Natl. Acad. Sci. USA 100, 13698-13703. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.