Abstract
The basis for segregation of sister chromosomes in bacteria is not established. We show here that two discrete ~150-kb regions, both located early in the right replichore, exhibit prolonged juxtaposition of sister loci, for 20 and 30 min, respectively, after replication. Flanking regions, meanwhile, separate. Thus, the two identified regions comprise specialized late-splitting intersister connections or snaps. Sister snap loci separate simultaneously in both snap regions, concomitant with a major global nucleoid reorganization that results in emergence of a bilobed nucleoid morphology. Split snap loci move rapidly apart to a separation distance comparable with one-half the length of the nucleoid. Concomitantly, at already split positions, sister loci undergo further separation to a comparable distance. The overall consequence of these and other effects is that thus far replicated sister chromosomes become spatially separated (individualized) into the two nucleoid lobes, while the terminus region (and likely, all unreplicated portions of the chromosome) moves to midcell. These and other findings imply that segregation of Escherichia coli sister chromosomes is not a smooth continuous process but involves at least one and likely, two major global transition(s). The presented patterns further suggest that accumulation of internal intranucleoid forces and constraining of these forces by snaps play central roles in global chromosome dynamics. They are consistent with and supportive of our previous proposals that individualization of sisters in E. coli is driven primarily by internally generated pushing forces and is directly analogous to sister individualization at the prophase to prometaphase transition of the eukaryotic cell cycle.
Keywords: E. coli chromosome, chromosome segregation, bacterial nucleoid
In bacteria, sister chromosomes segregate concomitant with DNA replication. We previously examined sister chromosome relationships over time in the Escherichia coli cell cycle (1). High temporal resolution (5–10 min) was achieved using synchronous populations (2). Sister relationships at individual loci were evaluated as in other studies. Also, uniquely, at a larger scale, whole nucleoid disposition and morphology were defined. This analysis identified a discrete transition, occurring part way through the replication process, in which the nucleoid becomes bilobed. This morphological change is accompanied by reciprocal repositioning of the replication origin (oriC) and terminus region (ter) and by strongly delayed splitting of one particular locus located near the replication origin (gln). These coordinate effects pointed to global reorganization of the nucleoid. We proposed that this reorganization resulted in spatial segregation of sister chromosomes and that it is analogous to the prophase to prometaphase transition of the eukaryotic cell cycle. We further proposed that, in both cases, sister individualization results from internally generated forces, more specifically, mechanical pushing effects (1, 3).
Other models for segregation of bacterial sister chromosomes through internal forces have been proposed, including (i) pulling apart of intersister connections because of stresses generated by longitudinal compaction, also suggested to be analogous to the mitotic prophase to prometaphase transition (4, 5), (ii) replisome pumping of plectonemic supercoils (6) pushing of sisters out from a replication factory (7), and thermally driven entropic segregation because of confinement within a cylindrical cell (8). For E. coli, internal force models match the fact that all molecules thus far implicated in sister segregation are known mediators of general basic DNA/chromosome structure (9).
A second group of bacterial chromosome segregation models invokes mechanisms in which sisters are separated by external motor-mediated pulling forces (e.g., analogous to anaphase segregation of sisters in eukaryotic organisms). Such forces are mostly suggested to be acted on discrete centromere-like regions. Three widely studied organisms, Caulobacter crescentus, Vibrio cholerae, and Bacillus subtilis, encode a Par system comprising a cis-acting locus (parS) and two proteins: ParB, which binds parS, and ParA, an ATPase that interacts with ParB and can polymerize on DNA (reviewed in ref. 10). The Par system is absent from E. coli and its proteobacterial γ-subdivision relatives (11). However, a centromere-like function is proposed for the E. coli migS locus (12, 13). In all cases, the primary centromere-like element is located near the replication origin. Motor-driven segregation focused on this element might then move sister origins to opposite poles (14, 15). Other proposed external pulling-type mechanisms invoke bacterial homologs of eukaryotic actin and tubulin, MreB and FtsZ, RNA polymerase (RNAP) action on origin-proximal genes biased in orientation away from oriC, or passive dispersal plus compaction (10, 16–18). Involvement of pulling mechanisms remains unclear, because, in several organisms, mutations in relevant determinants either do not affect segregation or perturb its regularity or completion rather than preventing it altogether (19–23). Thus, it was recently suggested for Caulobacter that Par-mediated anaphase-like effects may be preceded by processes related to preanaphase stages of the eukaryotic chromosomal program (23), similar to suggestions for E. coli (above).
Another issue for bacteria chromosome dynamics is whether sister chromosomes are subject to specific cohesion mechanisms. In E. coli, analysis of loci tagged with fluorescent proteins reveals that, at most positions throughout the genome, several minutes elapse between the time of replication and the time that two spatially resolved foci appear (24, 25) (Results). It is proposed that sisters are linked through topological intertwinings arising behind the replication fork (precatenanes), becoming separated as the fork moves forward (26). Another conundrum is how to reconcile progressive sister separation with sister separation through an abrupt global transition (above).
The current study explores these issues by further analyzing DNA replication, sister segregation, and global nucleoid dynamics in E. coli over time in the cell cycle.
Results
Replication Timing.
Under standard slow-growth conditions (1) (SI Materials and Methods), cells grow by a linear ~125-min cell cycle. Cell division is followed, after a short delay, by initiation and completion of DNA replication, which takes about 1 h, with concomitant separation of sister chromosomes followed by an additional period of about 1 h that includes finalization of sister separation in the terminus region and finally, the next cell division.
E. coli replicates its chromosome bidirectionally from a unique origin (oriC). Precise timing of replication initiation and progression around the chromosome, relative to cell birth, was defined in exponential cells. Relative population average abundances of sequences at 14 loci (Fig. 1A) were determined by quantitative real-time PCR (qPCR) and converted to absolute abundances by normalization to the number of oriC copies determined by flow cytometry (SI Materials and Methods and Fig. S1 A and B). For every distance from oriC, sequences of the left replichore are more abundant than sequences of the right replichore (Fig. 1B). Thus, left replichore replication precedes right replichore replication, as in other E. coli strains (27) (Fig. S1 B, D, and F). After accounting for the distribution of different cell ages in an exponential culture, these data define absolute times of replication after birth (Fig. 1C and SI Materials and Methods). The left replichore fork is ~7 min ahead of the right replichore fork, similar to prior measurements (27); replication speed is ~40 kb/min (38 ± 4), and the entire genome is replicated in ~60 min (61 ± 7).
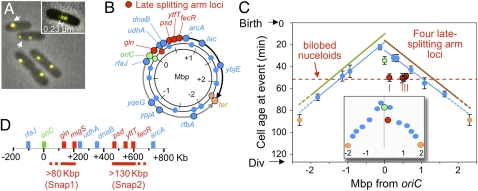
Fig. 1.
Left replichore replication precedes right replichore replication by ~7 min. (A) Chromosomal positions of 15 loci assayed by qPCR. (B) Copies per cell for each locus in exponential cell populations (three independent experiments ± 1 SD). (C) Replication times relative to cell birth (details in SI Materials and Methods and Fig. S1).
Sister Splitting Times Define Two Late-Splitting Intersister Snap Regions.
In fluorescent repressor operator system analysis (FROS), a locus tagged by a tet operator array bound by a cognate fluorescently tagged repressor (TetR-GFP) presents a single focus, if not yet replicated, or if replicated, with sister loci too close together to be distinguished (here, 230 nm) (Fig. 2A). Transit to the two-focus state operationally defines sister splitting. Splitting times were determined for 15 loci (Fig. 2B, outer ring) by analysis of exponential cells, taking into account relative abundances of cells at different stages (above) and then, comparing with predicted replication times defined above (Fig. 2C). Timing of bilobed nucleoid appearance was determined analogously (Fig. 2C, red dashed line).
Fig. 2.
Timing of sister splitting vs. replication. (A) FROS images. (Inset) Minimum detectable intersister distance. (B) Chromosomal positions of tetO array insertions assayed here (outer ring) or by Nielsen et al. (24) (middle ring). Late-splitting loci in red. (C) Sister separation times determined in exponential cells (two or three independent experiments ± 1 SD for each locus). The dashed red line shows analogously determined time of appearance of bilobed nucleoids. Regression lines for sites exhibiting normally delayed separation (blue) and left and right replichore replication (green and brown, respectively; from Fig. 1). Four vertical red lines point to four late-splitting loci. C Inset shows separation timing as defined from Nielsen et al. (24). (D) Positions of Snap1 and Snap2 along the right replichore.
Four left replichore loci and five right replichore loci exhibit splitting 7–10 min after they are replicated (Fig. 2C, blue). The same pattern, albeit with less precise definition of replication timing, was seen previously for multiple loci in both replichores (information in ref. 24 is reproduced in Fig. 2B, middle ring and Fig. 2C Inset) (25). This pattern represents the typical behavior of sister loci. Loci involved in initiation and termination of replication, oriC and ter, exhibit longer splitting delays, as seen previously (1, 19, 24, 25, 28).
We previously showed by FISH that the gln locus, located 130 kb distal to oriC in the right replichore (Fig. 2B, red), splits with an even greater delay after its predicted replication time than either oriC or ter (1). Furthermore, splitting at gln occurs in tight temporal linkage with appearance of bilobed nucleoids (1). These patterns are confirmed here (Fig. 2C, red circle and dotted line, respectively, and Discussion). Nielsen et al. (24) defined another late-splitting site, migS, located 80 kb farther downstream of gln (Fig. 2B, middle ring and red), whose splitting timing is very similar to that of gln (Fig. 2C Inset, red).
The present study further identifies three additional loci that exhibit strongly delayed splitting compared with other loci. These loci, psd, yftT, and fecR, lie in a contiguous ~130-kb segment located ~250 kb further along the right replichore from gln/migS (Fig. 2 B, red and D). Splitting at these loci is remarkable in several respects. (i) Sisters split at the same time at all three loci. (ii) The three loci also split at the same time as gln (and apparently, migS) (Fig. 2C, red), although the most ori-proximal and most ori-distal loci (gln and fecR, respectively) are separated by 0.5 Mb in genomic distance and 12 min in time of replication (Fig. 2 B–D). (iii) Loci exhibiting the typical 7–10 min delay in splitting occur just proximal to gln-migS (oriC) between gln-migS and psd-yftT-fecR (dnaB/udhA) and immediately distal to fecR (arcA). Moreover, these typical loci split before the very late-splitting loci (Fig. 2C). Thus, in the period immediately preceding their splitting, the gln-migS and psd-yftT-fecR regions comprise two discrete late-splitting intersister snaps: positions at which sisters maintain close juxtaposition while at the same time, sisters in flanking regions and the intervening ~200-kb region are separated (Figs. 2D and 3A; hereafter, Snap1 and Snap2). (iv) Temporally coordinated splitting of Snap1 and Snap2 is concomitant with appearance of bilobed nucleoids (Fig. 2C, dashed red line) and thus, the accompanying global nucleoid transition (1) (Discussion). These events occur when replication has proceeded approximately halfway around the chromosome in both directions (Fig. 2C).
Fig. 3.
Coordinate splitting at snap loci in synchronous populations (n = 100 per time point). (A) FISH image (cell at t = 30 min); map of FISH probe positions. (B) Appearance of cells exhibiting two foci over time in one cell cycle (Left). Cumulative curves defining segregation times (Right). Data for gln (gray) from Bates and Kleckner (1). (C) Per cell analysis of splitting at dnaB, arcA, psd, and fecR. Minority combinations (most excluded for clarity) represent <6% of cells at all time points.
Analogous analyses by FISH reveal splitting delays of 7–10 min for nonsnap loci and 20–30 min for snap loci (Fig. S2A). Delayed splitting at gln and psd was also observed in another strain background (Fig. S3). Fluorescent foci were detected at 95% efficiency by FROS and >90% efficiency by FISH (SI Materials and Methods).
Snap Splitting in Synchronous Populations.
We previously defined E. coli chromosome dynamics at high temporal resolution in synchronous cultures in the same conditions used here (1, 2). Cells tethered to a glass bead column release newly divided (newborn) cells. Aliquots of eluted cells collected over 5 min provide synchronous cultures that are then examined for events of interest over time after birth. Degree of synchrony is given by the slopes with which cells progress through basic stages (e.g., cell division) (Fig. 3B Right, gray dashes). Events separated in time by ≥5 min are resolved.
Synchronous populations were used to analyze sister-splitting status simultaneously by four-color FISH at four different loci: two Snap2 loci (psd and fecR) and immediately proximal and distal nonsnap loci dnaB and arcA (Fig. 3). In accordance with exponential cell analysis (Fig. 2C), two-focus cells appear first for dnaB, next for arcA, and then, simultaneously, for psd and fecR (Fig. 3C). Cumulative curve analysis of these data yields the percentage of cells that have progressed into or beyond the stage of interest as a function of time (Fig. 3B Right). Cumulative curves for psd and fecR are closely overlapping with one another and the curve for splitting at gln, as defined previously, confirming that Snap2 loci and the Snap1 locus gln split at the same time relative to cell birth (within 5 min of one another). Intriguingly, cumulative curves slopes for separation at nonsnap loci are less steep than for separation at psd and fecR, and the latter is the same as for cell division. Thus, snap locus segregation is apparently tightly linked to the cell division cycle, as shown previously (1, 2), whereas separation at nonsnap loci is less so. FROS analysis in synchronous populations gives the same timing as FISH for several loci (Fig. S2A).
These data permit per-cell analysis of splitting. Cells with splitting at dnaB only appear just before cells with splitting at dnaB and arcA only, which appear just before cells with splitting at all four loci (Fig. 3C). Importantly, >90% of cells exhibited splitting at both Snap2 loci or neither Snap2 locus; cells exhibiting splitting at only one of the two loci occur only at the very low background level seen for other noncanonical combinations. Thus, Snap2 loci psd and fecR split contemporaneously in individual cells.
Abrupt Long-Distance Separation of Snap Loci and Correlated Further Separation of Already Split Nonsnap Loci.
Synchronous cell populations were analyzed for the distance between sister foci as a function of time after birth at snap loci and nearby nonsnap loci that separate before snap splitting.
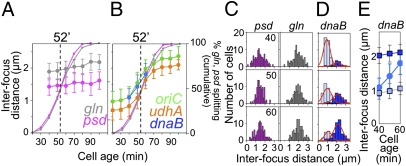
Cells with split snap loci appear at t = 40, 50, and 60 min, comprising 15%, 40%, and 70% of cells (Figs. 3B and 4 A and B). The population average distance between sister foci is the same at all three time points, 2.0 (gln) and 1.5 μm (psd) (Fig. 4A and Fig. S2 B and C). Intersister distances comprise symmetrical distributions centered on these values, with no obvious shoulder of shorter distances (Fig. 4C and Fig. S2C). These data suggest that snap loci move to final widely separated positions relatively abruptly (on time scales less than intertime point intervals; ≤10 min). Slow progressive sister separation would have yielded progressively increasing average intersister distances at the three time points and a prominent fraction of cells with closer-spaced sisters. Because nucleoid lengths during the splitting period are 2.5–3.0 μm (Fig. S4C), sister loci move abruptly far apart to distances corresponding to one-half the length of the nucleoid or more. Interestingly, although one-half of the chromosome remains to be replicated, intersister distances increase little after splitting, implying that a new relatively stable configuration has been achieved.
Fig. 4.
Intersister distances increase abruptly. For each indicated locus, two-focus cells in synchronous populations were measured for distances between sister foci at indicated times after birth (n = 100 per time point). (A and B) Cells undergo snap splitting at t = 40–60 min. (Cumulative curves from Fig. 5; 50% at t = 52). (A and C) Snap loci gln and psd show immediate wide intersister separation (A; average ±1 SD) and a uniform intersister distance distribution (C). (B, D, and E) Concomitant with snap splitting, at nonsnap loci (oriC, udhA, and dnaB), already separated sisters exhibit immediate further separation to a wide distance. (B) Average intersister distances increase progressively as cells undergo snap splitting. (D) Cells not yet split at either of two snap loci psd and fecR (light blue) and cells split at both snap loci (dark blue) exhibit distinct intersister distance distributions. (E) Average intersister distance increases progressively as the weighted average contributions of the two populations in D. (A–C) FROS analysis (Fig. 2B). (D and E) FISH analysis of Fig. 3.
In temporal correlation with loss of sister colocalization at snaps, loci that have undergone splitting previously undergo further sister separation. Similarly to separation at snap loci, sisters are immediately placed at separation distances of one-half the nucleoid length or more, with little further increase thereafter. As shown previously (1), sister oriCs initially separate at approximately t = 30 min and then, concomitant with the appearance of bilobed nucleoids and splitting at gln, abruptly move far apart, with little further separation thereafter. Correspondingly, average intersister oriC distances increase, in parallel with appearance of cells containing split snaps, as more and more cells undergo the snap-splitting transition and then, further increase only slowly (Fig. 4B, green). The same pattern occurs at other loci. At udhA, intersister distance increases from 0.7 to 1.7 μm at t = 40–60 min (orange). In four-color FISH analysis (Fig. 3), cells with splitting at two Snap2 loci (fecR and psd) or neither Snap2 locus show larger and smaller intersister distances for previously split foci of dnaB, ~2.2 and ~1 μm, respectively (Fig. 4 D and E), with average dnaB intersister distance increasing over time at t = 40–60 min (Fig. 4 B and E). Bimodal distribution of intersister distances for already split dnaB underscores the significance of unimodal distributions for snap loci.
Bilobed Nucleoids Represent Long-Distance Separation of Sister Chromosomes.
Part of the way through the cell cycle, the nucleoid changes morphology from unilobed to bilobed state, as defined in fixed cells stained with DNA-intercalating dye DAPI (0.5 μg/mL) (1) (Fig. 5A, also in MG1655 strain background). Bilobed nucleoids appear contemporaneously with splitting at snap loci (as seen by exponential cell and cumulative curve analyses) (Figs. 2C and 5B) (1). Splitting is also simultaneous within individual cells: 90% of unilobed nucleoids exhibit a single gln or psd focus and 90% of bilobed nucleoids exhibit split psd or fecR loci; conversely, 90% of cells with a single focus at either or both snap loci are unilobed, whereas 90% of cells with splitting at either or both loci are bilobed (Fig. 5C). Approximately ten percent of exceptional cases may reflect absence of perfect per-cell synchrony and/or dynamic fluctuations of the nucleoid during the transition.
Fig. 5.
Sister separation at snap loci is concomitant with nucleoid expansion and sister individualization. (A) DAPI images from synchronized cells before (0–40 min) and after (60–100 min) nucleoid splitting. (B) Fraction of cells containing a bilobed nucleoid over time and corresponding cumulative curve (cyan symbols; 50% at t = 52 min) superimposed on cumulative curves for separation of three snap loci (orange, magenta, and gray lines from Fig. 3B). (C) Correlations among nucleoid status, sister separation at snap loci gln or psd (FROS), and cell length. (D) Cells with bilobed nucleoids usually have one sister locus in each lobe defined for asynchronous FROS-marked cells (Fig. 2B) (n = 100 per strain).
Because bilobed nucleoid appearance is accompanied by separation of snap and nonsnap loci to distances of one-half or more the length of the cell (above), each nucleoid lobe might represent one sister chromosome. To test this possibility, cells with bilobed nucleoids were examined for sister focus positions. Exponential cells exhibiting a bilobed nucleoid were analyzed by FROS at each of five loci: oriC, Snap1 locus gln, and nonsnap loci in the left (rfaJ) and right (dnaB and udhA) replichores. At all five loci, >90% of cells containing both bilobed nucleoids and split sisters exhibit one sister in each lobe (Fig. 5D) for cells of the length predominant at the time of sister splitting (3–4 μm) (1), cells where splitting occurred at a shorter length (2–3 μm), and longer (older) cells (4–5 μm). Thus, the transition at which snap loci split and bilobed nucleoids appear results in the spatial separation of thus far replicated sister chromosomes into the two lobes (i.e., sister individualization). Interestingly, also, appearance of bilobed nucleoids is accompanied by a discrete increase in nucleoid volume in accordance with increased spatial separation of sister chromosomes (Fig. S2 D and E).
Two Sequential Discrete Transitions Reflect Underlying Nucleoid Asymmetry.
Previous synchronous cell analysis described oriC and ter positions and the boundaries of the nucleoid(s) and cell over time in the cell cycle (1) (Fig. 6 and Fig. S4). Sister oriCs move out to opposite ends of the cell sequentially in two discrete transitions: t = 35 min (T1) and t = 55 min (T2) (Fig. 6). Movement of sister oriC is asymmetric, with a specific polarity relative to which side of the nucleoid contains ter. At T1, one sister oriC moves to the ter-distal end of the nucleoid, whereas its sibling remains near midcell. At T2, the midcell-localized sibling moves in the opposite direction to the ter-proximal end of the nucleoid. Interestingly, both transitions place the moving oriC at the same specific distance from both the end of the nucleoid and the end of the cell, and both distances remain constant thereafter until the end of the cell cycle (Fig. 6 and Fig. S4). oriC positioning may, thus, be determined relative to the end of the cell or with respect to the nucleoid through internally determined positioning.
Fig. 6.
Dynamic changes of oriC, ter, and the nucleoid through the cell cycle. Population average distances from midcell of oriC, ter, and the ends of the nucleoid and cell (adapted from ref. 1).
T2 is the transition that gives bilobed nucleoids, with the accompanying changes described above. During this same transition, ter moves in to midcell, effectively switching positions with the oriC in the same (ter) side of the cell (Fig. 6). T1 has no obvious morphological effect on the nucleoid but shares other analogies with T2. T1 and T2 both deposit an oriC at its final position (above). T1 involves the abrupt initial separation of sister oriCs to a distance of about one-half the nucleoid length (1.0 ± 0.2 μm vs. 2.3 ± 0.4 μm), similarly to the observed abrupt further separation of sister oriCs at T2 (above). T1 oriC separation distance far exceeds that required for resolution of two discrete sister foci (230 nm; above), excluding the progressive drifting apart of sister foci to a spatially resolvable separation distance. Perhaps T1 involves release of intersister snaps at oriC or analogous features. Asymmetric sequential positioning of oriC at T1 and T2 and asymmetric nucleoid morphology at T2 may both reflect underlying intrinsic nucleoid asymmetry.
Discussion
Above results define E. coli sister segregation dynamics at individual loci in relation to whole nucleoid morphogenesis. (i) At most loci around the chromosome, sisters are closely juxtaposed for 7–10 min after replication. (ii) Superimposed on this progressive process is a prominent, abrupt global reorganization that involves movement throughout the nucleoid, including replicated regions and unreplicated ter, and results in the appearance of bilobed nucleoids, with thus far replicated sister chromosomes spatially separated into the two lobes. (iii) At two unique regions, both located early in the right replichore, sisters remain closely juxtaposed much longer than at other loci, thus comprising late-splitting intersister snaps. Sister splitting in these two snap regions then occurs, synchronously, as part of the overall nucleoid reorganization. The same sister separation patterns are seen by FISH and FROS, removing any possibility that methodology influences experimental results.
Late-Splitting Intersister Snaps.
Late-splitting loci define two discrete regions of ~100–150 kb that comprise late-splitting intersister snaps. Both snap regions lie in the first one-third of the right replichore for reasons to be determined. Genomic analyses do not identify any feature that correlates with snaps (e.g., base composition or mRNA expression profiles) (Fig. S5) or binding sites for known abundant chromosomal proteins including DnaA, SeqA, IHF, and Fis. However, a cluster of four ribosomal RNA genes occurs just distal to oriC on the right chromosome arm, raising the possibility of a relationship between highly transcribed regions and snap dynamics. The molecular basis for prolonged maintenance of sister colocalization in snap regions, thus, remains to be determined. Direct DNA/DNA pairing (29) and intranucleoid pressure (ref. 6, below) are possibilities. Snap1 and Snap2 may be the only late-splitting regions in interstitial (nonoriC and nonter) positions around the chromosome. Among more than 20 loci examined in this and previous studies, no other such regions were detected. We note that lac splits concomitant with splitting at gln and appearance of bilobed nucleoids (1); however, this is presumably only because of its position along the chromosome, which, by chance, dictates sister segregation at the time of the snap-mediated global transition (Fig. 2C).
What Keeps Sister Chromosomes Close Together for ~7–10 Min?
Temporal analysis (above) implies that, by the time a locus detectably splits, the replication fork from which it emerged will have progressed another 300–400 kb along the chromosome. That is, each replication fork is trailed by a 300- to 400-kb sliding window of sister juxtaposition. Sister loci might remain spatially colocalized, because they are loosely linked through precatenanes (in the Introduction). However, alternative scenarios also merit consideration, particularly given identification of long-lived juxtaposition in snap regions (above).
Wide Separation of Sister Chromosomes Through a Discrete Global Transition.
Most considerations of how sister chromosomes segregate over long distances have envisioned a smooth, continuous process in which sister loci move steadily apart. We show here, confirming previous indications (1), that sister segregation in E. coli involves at least one and probably, two discrete, noncontinuous transitions. The most prominent transition effects the spatial separation of sister chromosomes, producing two morphologically discrete nucleoid lobes, with an accompanying increase in total nucleoid volume. Concomitantly, changes in disposition occur in synchrony throughout the nucleoid. (i) Two intersister snap regions undergo abrupt splitting and rapid wide separation of sister loci, whereas concomitantly, in loci that have already undergone sister splitting, sisters undergo an abrupt wider separation (Fig. 7A). (ii) ter, which has not yet been replicated, moves abruptly to midcell, opposite to poleward movement of the closer sister oriC. All presented findings suggest that there is a discrete, relatively rapid (<10 min) transition from one state to another.
Fig. 7.
Sister dynamics. (A) Coordinate loss of links at Snap1 and Snap2 results in wide separation of thus far replicated sister chromosomes (solid and dotted black lines). (B) Proposed two-phase segregation of sisters asymmetrically with respect to the mother nucleoid (red/blue and green, respectively).
Source(s) of Segregation Force(s).
We infer that the separation of sisters at discrete ~150-kb intersister snap regions is required for wide separation of sisters into distinct nucleoid lobes and thus, overall nucleoid reorganization. Such separation could reflect either pulling or pushing forces.
Sister chromosomes could be pulled farther and farther apart to the poles. Snap regions would resist this tendency but would finally also be pulled apart, with concomitant rapid retraction of sister snap loci to widely separated positions. Pulling could result ultimately from external forces exerted on centromere-like regions anchored at/near the poles and/or multiple positions within the nucleoid. In either case, such forces must effect the segregation of entire whole chromosomal domains rather than pulling out the chromosome fibers immediately adjacent to the site of imposed pulling forces. Recent indications that the E. coli nucleoid is very stiff (6) support the possibility of global physical coherence. Alternatively, anchored focal points (e.g., centromere-like elements) might nucleate compaction that spreads progressively out, drawing in other regions.
Forces arising internally within the nucleoid could tend to push sisters apart. Snaps would prevent such forces from effecting full sister segregation. When the level of force accumulates above a critical level exceeding the strength of the snaps, these links would be released, permitting sister chromosomes to abruptly move apart from one another into separate spaces as physically coherent units (6).
We favor pushing models, because the necessary forces can be generated without specialized motors, molecular apparatus, or organism-specific features (in the Introduction). Also, our analysis suggests that sister oriCs are positioned as a consequence of the two discrete transitions rather than a prerequisite, suggesting that oriC is not serving as an anchor point for ensuing compaction-mediated pulling. In fact, by a pushing scenario, internal forces would propel the oriCs out to the ends of the cell. Such an effect could explain the release of anchoring and ensuing polar localization of origin(s) seen in other types of bacteria (23).
Appearance of Bilobed Nucleoids Comprises Commitment to Sister Segregation.
When bilobed nucleoids appear at already replicated loci, one sister locus occurs in each nucleoid lobe (Fig. 5D). Sister material created subsequently will likely move directly into the two sister domains. Thus, sister individualization into bilobed nucleoids comprises commitment of the nucleoid to ultimately achieving full segregation.
Arrival at this committed state seems to involve two discrete transitions (T1 and T2; above). Both involve asymmetric movements of oriC and ter and culminate, after T2, in a spatially asymmetric configuration with one large and one small nucleoid lobe. We propose that the smaller lobe comprises material of one sister, whereas the large lobe comprises the other sister plus unreplicated mother DNA (Fig. 7B). This is because (i) the small lobe contains one oriC, whereas the large lobe contains the other oriC plus unreplicated ter (Fig. 6 and Fig. S4), and (ii) the larger lobe does not change in size for the rest of the cell cycle (Fig. S4C), at the end of which it must contain one complete sister chromosome (i.e., one genome equivalent). In the bilobed nucleoid stage, one sister plus remaining mother material would also comprise one genome equivalent. As replication proceeds, material would move from the mother to one sister within the larger lobe, with no change in total genome content (size), whereas material of the other sister would accumulate progressively in the smaller lobe (Fig. 7B). We further propose that sister segregation occurs in two stages: one sister moves out to one end of the cell at T1 and the other sister moves out to the other end of the cell at T2, with concomitant inward movement of the mother nucleoid (Fig. 7B). Asymmetric sequential segregation of sisters also fits with the fact that sister positioning is correlated with leading/lagging strand replication fork bias (30).
Spatial asymmetry in oriC and ter dynamics plus corresponding asymmetry in nucleoid morphology point to existence of internal asymmetry within the nucleoid per se.
Analogies with the Eukaryotic Prophase to Prometaphase Transition.
During the eukaryotic cell cycle, long, thin prophase chromosomes, with sisters comprising a single morphological unit, become shorter, fatter prometaphase chromosomes with sisters as individualized side by side units (1, 3). This transition can be accompanied by increased chromosome volume (3). Also, during meiotic prophase, sisters cycle reversibly between single and individualized chromatin states, with concomitant chromatin volume variation (3, 31). We proposed previously that these phenomena reflect the existence of internal pushing forces, which push sister chromatin masses apart (3), and that the intersister pushing forces responsible for individualization of sisters in E. coli should be analogous to those that promote these effects during the eukaryotic cell cycle (1). The above findings are consistent with, and supportive of, these ideas.
Materials and Methods
Bacterial Strains.
FISH analysis used NK9386; FROS experiments used NK9386 derivatives carrying tetO arrays (SI Materials and Methods and Table S1).
Cell Culture Preparation.
For exponential cell analysis, cells were serially diluted and grown overnight to OD0.2 at 450 nm before assaying. Synchronous populations were obtained as in refs. 1 and 2. Individual 5-min samples were collected, placed into shaking culture for varying amounts of time, and then, assayed immediately (FROS) or after fixation (FISH and DAPI) (SI Materials and Methods). Adverse effects of high TetR-YFP expression were avoided (SI Materials and Methods).
Fluorescence Imaging.
FROS imaging was performed on living cells after 1 h of 0.02% arabinose (TetR-GFP) induction. Cells were applied to prewarmed agarose-coated slides (1% in AB minimal medium), covered with a slip, and immediately imaged in a temperature-controlled environment at 30 °C. For FISH, cells were fixed in 2.5% paraformaldehyde and processed as in ref. 1; 3-kb probes were amplified from genomic DNA (Table S2) and chemically labeled (SI Materials and Methods). Multicolor fluorescent beads on slides were used to align independent color channels and calibrate DAPI staining. All images were acquired with a Zeiss AxioImager Z1 microscope plus Hamamatsu Electron Multiplier charge-coupled device (CCD) camera.
Acknowledgments
We thank Mara Prentiss and Beth Weiner for insightful comments and Beth Weiner for manuscript preparation. Research was supported by National Institutes of Health Grants GM025326 (to N.K.) and GM20627 (to D.B.) and Baylor College of Medicine (D.B.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1019593108/-/DCSupplemental.
References
- 1.Bates D, Kleckner N. Chromosome and replisome dynamics in E. coli: Loss of sister cohesion triggers global chromosome movement and mediates chromosome segregation. Cell. 2005;121:899–911. doi: 10.1016/j.cell.2005.04.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bates D, et al. The Escherichia coli baby cell column: A novel cell synchronization method provides new insight into the bacterial cell cycle. Mol Microbiol. 2005;57:380–391. doi: 10.1111/j.1365-2958.2005.04693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kleckner N, et al. A mechanical basis for chromosome function. Proc Natl Acad Sci USA. 2004;101:12592–12597. doi: 10.1073/pnas.0402724101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holmes VF, Cozzarelli NR. Closing the ring: Links between SMC proteins and chromosome partitioning, condensation, and supercoiling. Proc Natl Acad Sci USA. 2000;97:1322–1324. doi: 10.1073/pnas.040576797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marko JF, Siggia ED. Polymer models of meiotic and mitotic chromosomes. Mol Biol Cell. 1997;8:2217–2231. doi: 10.1091/mbc.8.11.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wiggins PA, Cheveralls KC, Martin JS, Lintner R, Kondev J. Strong intranucleoid interactions organize the Escherichia coli chromosome into a nucleoid filament. Proc Natl Acad Sci USA. 2010;107:4991–4995. doi: 10.1073/pnas.0912062107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lemon KP, Grossman AD. The extrusion-capture model for chromosome partitioning in bacteria. Genes Dev. 2001;15:2031–2041. doi: 10.1101/gad.913301. [DOI] [PubMed] [Google Scholar]
- 8.Jun S, Mulder B. Entropy-driven spatial organization of highly confined polymers: Lessons for the bacterial chromosome. Proc Natl Acad Sci USA. 2006;103:12388–12393. doi: 10.1073/pnas.0605305103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Reyes-Lamothe R, Wang X, Sherratt D. Escherichia coli and its chromosome. Trends Microbiol. 2008;16:238–245. doi: 10.1016/j.tim.2008.02.003. [DOI] [PubMed] [Google Scholar]
- 10.Gerdes K, Howard M, Szardenings F. Pushing and pulling in prokaryotic DNA segregation. Cell. 2010;141:927–942. doi: 10.1016/j.cell.2010.05.033. [DOI] [PubMed] [Google Scholar]
- 11.Yamaichi Y, Niki H. Active segregation by the Bacillus subtilis partitioning system in Escherichia coli. Proc Natl Acad Sci USA. 2000;97:14656–14661. doi: 10.1073/pnas.97.26.14656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamaichi Y, Niki H. migS, a cis-acting site that affects bipolar positioning of oriC on the Escherichia coli chromosome. EMBO J. 2004;23:221–233. doi: 10.1038/sj.emboj.7600028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fekete RA, Chattoraj DK. A cis-acting sequence involved in chromosome segregation in Escherichia coli. Mol Microbiol. 2005;55:175–183. doi: 10.1111/j.1365-2958.2004.04392.x. [DOI] [PubMed] [Google Scholar]
- 14.Toro E, Shapiro L. Bacterial chromosome organization and segregation. Cold Spring Harb Perspect Biol. 2010;2:a000349. doi: 10.1101/cshperspect.a000349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schofield WB, Lim HC, Jacobs-Wagner C. Cell cycle coordination and regulation of bacterial chromosome segregation dynamics by polarly localized proteins. EMBO J. 2010;29:3068–3081. doi: 10.1038/emboj.2010.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dworkin J, Losick R. Does RNA polymerase help drive chromosome segregation in bacteria? Proc Natl Acad Sci USA. 2002;99:14089–14094. doi: 10.1073/pnas.182539899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dye NA, Shapiro L. The push and pull of the bacterial cytoskeleton. Trends Cell Biol. 2007;17:239–245. doi: 10.1016/j.tcb.2007.03.005. [DOI] [PubMed] [Google Scholar]
- 18.Norris V, Woldringh C, Mileykovskaya E. A hypothesis to explain division site selection in Escherichia coli by combining nucleoid occlusion and Min. FEBS Lett. 2004;561:3–10. doi: 10.1016/S0014-5793(04)00135-8. [DOI] [PubMed] [Google Scholar]
- 19.Wang X, Sherratt DJ. Independent segregation of the two arms of the Escherichia coli ori region requires neither RNA synthesis nor MreB dynamics. J Bacteriol. 2010;192:6143–6153. doi: 10.1128/JB.00861-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saint-Dic D, Frushour BP, Kehrl JH, Kahng LS. A parA homolog selectively influences positioning of the large chromosome origin in Vibrio cholerae. J Bacteriol. 2006;188:5626–5631. doi: 10.1128/JB.00250-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yamaichi Y, Fogel MA, Waldor MK. par genes and the pathology of chromosome loss in Vibrio cholerae. Proc Natl Acad Sci USA. 2007;104:630–635. doi: 10.1073/pnas.0608341104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Toro E, Hong SH, McAdams HH, Shapiro L. Caulobacter requires a dedicated mechanism to initiate chromosome segregation. Proc Natl Acad Sci USA. 2008;105:15435–15440. doi: 10.1073/pnas.0807448105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shebelut CW, Guberman JM, van Teeffelen S, Yakhnina AA, Gitai Z. Caulobacter chromosome segregation is an ordered multistep process. Proc Natl Acad Sci USA. 2010;107:14194–14198. doi: 10.1073/pnas.1005274107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nielsen HJ, Li Y, Youngren B, Hansen FG, Austin S. Progressive segregation of the Escherichia coli chromosome. Mol Microbiol. 2006;61:383–393. doi: 10.1111/j.1365-2958.2006.05245.x. [DOI] [PubMed] [Google Scholar]
- 25.Espeli O, Mercier R, Boccard F. DNA dynamics vary according to macrodomain topography in the E. coli chromosome. Mol Microbiol. 2008;68:1418–1427. doi: 10.1111/j.1365-2958.2008.06239.x. [DOI] [PubMed] [Google Scholar]
- 26.Wang X, Reyes-Lamothe R, Sherratt DJ. Modulation of Escherichia coli sister chromosome cohesion by topoisomerase IV. Genes Dev. 2008;22:2426–2433. doi: 10.1101/gad.487508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Breier AM, Weier HU, Cozzarelli NR. Independence of replisomes in Escherichia coli chromosomal replication. Proc Natl Acad Sci USA. 2005;102:3942–3947. doi: 10.1073/pnas.0500812102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y, Sergueev K, Austin S. The segregation of the Escherichia coli origin and terminus of replication. Mol Microbiol. 2002;46:985–996. doi: 10.1046/j.1365-2958.2002.03234.x. [DOI] [PubMed] [Google Scholar]
- 29.Danilowicz C, et al. Single molecule detection of direct, homologous, DNA/DNA pairing. Proc Natl Acad Sci USA. 2009;106:19824–19829. doi: 10.1073/pnas.0911214106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.White MA, Eykelenboom JK, Lopez-Vernaza MA, Wilson E, Leach DR. Non-random segregation of sister chromosomes in Escherichia coli. Nature. 2008;455:1248–1250. doi: 10.1038/nature07282. [DOI] [PubMed] [Google Scholar]
- 31.Dawe RK, Sedat JW, Agard DA, Cande WZ. Meiotic chromosome pairing in maize is associated with a novel chromatin organization. Cell. 1994;76:901–912. doi: 10.1016/0092-8674(94)90364-6. [DOI] [PubMed] [Google Scholar]