Abstract
Telomeres consist of an elaborate, higher-order DNA architecture, and a suite of proteins that provide protection for the chromosome terminus by blocking inappropriate recombination and nucleolytic attack. In addition, telomeres facilitate telomeric DNA replication by physical interactions with telomerase and the lagging strand replication machinery. The prevailing view has been that two distinct telomere capping complexes evolved, shelterin in vertebrates and a trimeric complex comprised of Cdc13, Stn1 and Ten1 (CST) in yeast. The recent discovery of a CST-like complex in plants and humans raises new questions about the composition of telomeres and their regulatory mechanisms in multicellular eukaryotes. In this review we discuss the evolving functions and interactions of CST components and their contributions to chromosome end protection and DNA replication.
Key words: telomere, telomerase, telomere protein, CTC1, STN1, TEN1, OB-fold, arabidopsis, DNA polymerase alpha, RPA
Telomere Protein Complexes: Shelterin versus CST
Vertebrate telomeres are bound by six telomere-specific proteins that assemble into a complex termed shelterin1 (Fig. 1A). The individual components, TRF1, TRF2, TIN2, Rap1, TPP1 and POT1, each play defined roles in telomere protection. These include limiting DNA degradation, preventing ATM and ATR-activation and inhibiting DNA repair activities such as non-homologous end joining or homology directed repair.2 TRF1 and TRF2 bind to the duplex region of the telomere while POT1 binds to the 3′ overhang on the G-rich strand. TIN2 and TPP1 form a bridge between TRF1/2 and POT1 linking the telomere duplex and the G-overhang.1 POT1 binds to the overhang via two adjacent oligonucleotide-oligosaccharide binding folds (OB-folds).3,4 Fission yeast telomeres also assemble with a shelterin-like complex that contains obvious orthologs of vertebrate TRF1/2 (Taz1), Rap1 and POT1.5 Although the vertebrate and fission yeast complexes differ in subunit arrangement, the overall structure seems quite similar as the S. pombe duplex binding protein Taz1 is linked to Pot1 and the G-overhang via a series of bridging proteins which include Rap1, Poz1, Tpz1 and Ccq1 (Fig. 1C). Tpz1 appears to be the functional homolog of TPP1.
Figure 1.
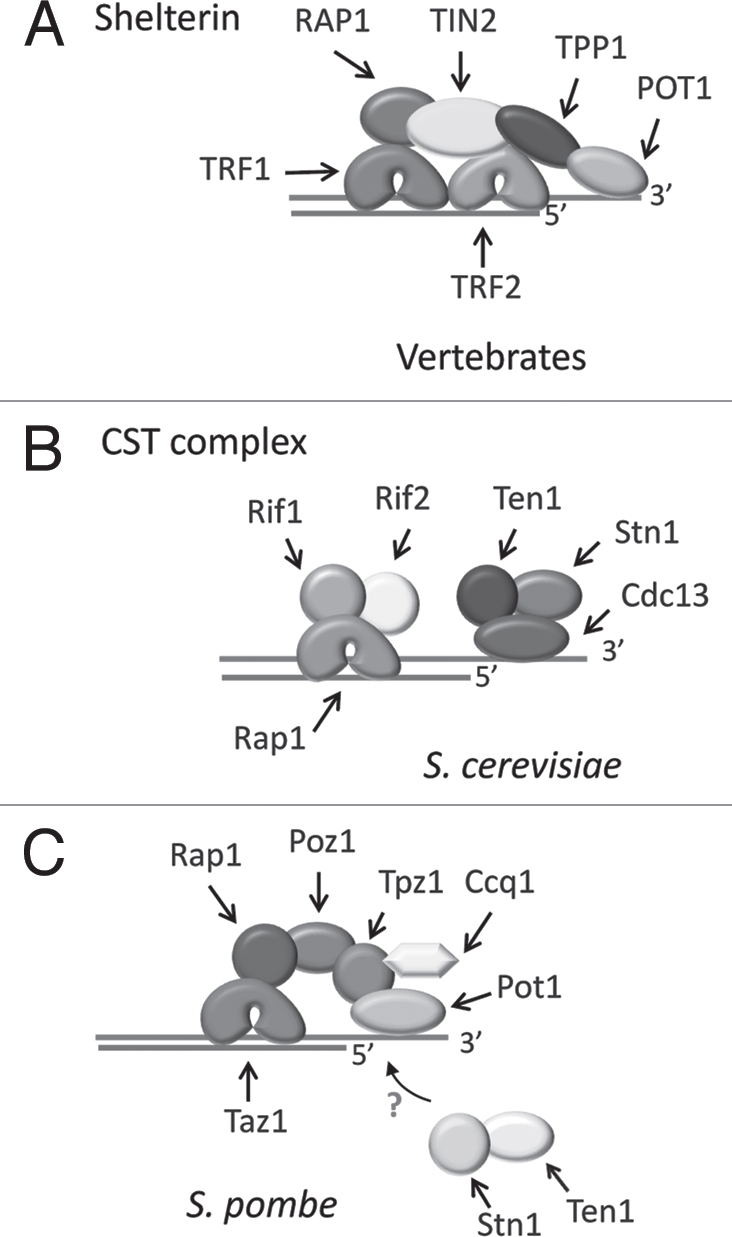
Telomere capping complexes in vertebrates and yeast. (A) The six-member shelterin complex associates with both single- and double-strand regions of the vertebrate telomeric DNA. (B) Budding yeast telomeres are protected by the trimeric Cdc13 Stn1 Ten1 (CST) complex, which assembles on the G-overhang. The duplex region of the telomere is bound by a separate complex containing Rap1, Rif1 and Rif2. (C) Fission yeast telomeres associate with a six member shelterin-like complex. In addition, Stn1 and Ten1 contribute to chromosome end protection, but it is not known how they interact with other telomere proteins.
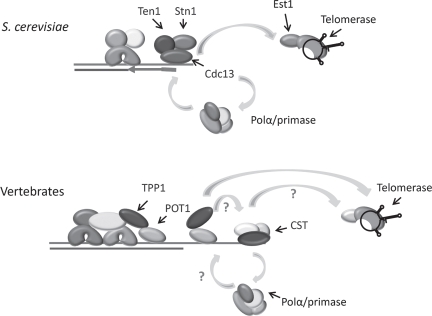
Intriguingly, budding yeast telomeres are not protected by a shelterin-like complex. Although the double-strand region of the telomere is bound by Rap1 and two associated factors, these proteins are not involved in chromosome end protection. Instead this function is fulfilled by a trimeric complex, CST, comprised of Cdc13, Stn1 and Ten1, which associates with the G-overhang (Fig. 1B).6 None of the CST components show obvious sequence identity to POT1, TPP1/Tpz1 or other shelterin constituents.4 S. cerevisiae CST plays a dual role in telomere protection and modulation of telomere replication.7 Although Cdc13 is the main DNA-binding subunit, all three proteins function in end-protection and removal of any subunit results in degradation of the telomeric C-strand, accumulation of long G-overhangs, activation of a DNA-damage response and a late S/G2 cell cycle arrest. Cdc13 and Stn1 play key roles in telomere replication.8–10 During late S/G2, phosphorylation of Cdc13 promotes a direct interaction between Cdc13 and the Est1 subunit of telomerase.11 This interaction enhances telomerase extension of the chromosome terminus.12,13 Subsequent dephosphorylation of Cdc13 limits telomerase action by reducing Est1 binding and increasing Stn1 binding.11,14 Cdc13 and Stn1 then appear to coordinate fill-in of the complementary C-strand by recruiting DNA Polα/primase through direct interactions with the Pol1 and Pol12 subunits of DNA Polα.15,16 Despite the lack of sequence similarity to POT1, the DNA binding domain of Cdc13 consists of an OB-fold that is structurally similar to the OB-folds in the DNA-binding domain of POT1.3,4,17 This discovery led to the idea that Cdc13 is the functional homolog of POT1 and further suggested that shelterin had replaced CST in vertebrate cells. This impression was reinforced when POT1 or TPP1 depletion was shown to cause a severe telomere uncapping phenotype analogous to that observed after removal of the S. cerevisiae CST complex.18–20 However, recent genetic and structural studies reveal that budding yeast CST is more closely related to Replication Protein A (RPA) than to POT1-TPP1.21,22
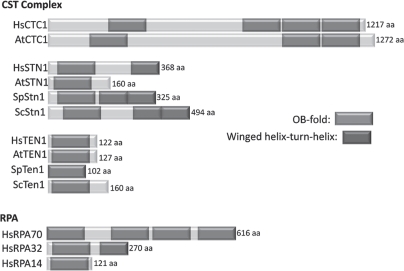
RPA is a heterotrimer that binds ssDNA through a series of OB-folds23 (Fig. 2). RPA70 contains four OB-folds, three of which contact DNA. RPA32 contains one OB-fold that also contacts DNA and a C-terminal winged helix (WH) protein interaction domain. RPA14 is comprised of a single OB-fold that is needed for complex formation. Protein structure prediction first suggested that Stn1 and Ten1 might contain OB-folds resembling those of RPA32 and RPA14.21 X-ray crystallography has since confirmed that budding yeast (C. tropicalis) Stn1-Ten1 and Rpa2-Rpa3 (RPA32-RPA14) complexes have substantial structural similarity in their OB-fold motifs, subunit interaction surfaces and in the Stn1 N- and C-terminal extension regions22 (Fig. 2). However, there are significant differences in the relative orientation of the subunits and in the structure of most of the connecting loop regions. Also, the C-terminal extension of Stn1 contains two WH motifs instead of the single motif found in Rpa2.22,24 Thus, while CST shares significant structural features with RPA, it is tailored to perform a different biological function.
Figure 2.
Similar domain structure in CST and RPA. The predicted domain structure is shown for CTC1 from humans and Arabidopsis and STN1 and TEN1 from humans, Arabidopsis, S. pombe and S. cerevisiae. For comparison, human RPA structure is shown.
Conservation of the CST Complex
The rapid evolution and resulting sequence divergence of telomere proteins makes it difficult to identify orthologs from other species using purely bioinformatics approaches.4 When database searches revealed potential Stn1 orthologs in a wide range of organisms,21,25 it was unclear whether they were bona-fide telomere proteins. The same cross-species database searches failed to identify orthologs of Ten1 and Cdc13. The potential Stn1 orthologs showed low sequence identity with budding yeast Stn1 (17.7% identity and 54.4% similarity for ScStn1 vs. Arabidopsis STN1; 21.5% identity and 59.5% similarity for ScStn1 vs. human STN1), but structure prediction programs revealed OB-fold domains similar to that found in RPA32 (Fig. 2). Subsequent disruption of the S. pombe STN1 gene demonstrated a role in telomere protection as the cells exhibited rapid telomere loss and end-to-end fusion of chromosomes.25 A tentative SpTen1 ortholog with a putative OB-fold was then identified by more sensitive bioinformatic analyses. TEN1 gene disruption gave the same telomere loss and end fusion phenotype as the STN1 disruption. The Ten1 protein was also shown to interact with Stn1 and to colocalize with Stn1 and Pot1 at telomeres.25 Interestingly, the Stn1-Ten1 complex does not appear to interact with Pot1, suggesting that fission yeast contain separate Stn1-Ten1 and Pot1/shelterin-like complexes that are both required for telomere protection. Thus far an ortholog of Cdc13 or the plant and vertebrate CTC1 remains to be identified in S. pombe.
Arabidopsis CST was uncovered through a combination of bioinformatic and genetic approaches.26,27 A putative STN1 ortholog was identified in the plant genome and the in vivo function was determined by analyzing the phenotype of STN1 null plants.26 Mutants showed profound defects in chromosome end protection and telomere maintenance (see below). Bioinformatics was also used to reveal a putative TEN1 ortholog based on similarity to human TEN1. As in yeast, AtSTN1 and AtTEN1 interact in vitro (Leehy K and Shippen D, unpublished data). CTC1 (Conserved Telomere maintenance Component 1) was identified using a genetic screen for mutations that cause telomere capping defects.27 CTC1 lacks sequence identity to any known gene but structure prediction programs (HHpred and Metaserver) indicate that the encoded protein contains multiple OB-folds with homology to the OB-folds from RPA70 (Fig. 2). CTC1 and STN1 interact in vitro and the phenotype of a CTC1 null plant is similar to that of a STN1 mutant or the CTC1/STN1 double mutant. Thus, CTC1 and STN1 appear to function in the same pathway for chromosome end protection and telomere maintenance in Arabidopsis.
The plant CTC1 sequence was employed in database searches using PSI-BLAST and HHpred to identify vertebrate CTC1.27 While the overall level of sequence identity was low (14% identity, 26% similarity between the human and Arabidopsis CTC1), the predicted secondary structure was similar throughout the length of the protein and the potential OB-folds again resembled those of RPA70 (Fig. 2). Subsequent siRNA knockdown of human CTC1 resulted in various telomere defects and genomic instability (see below). Mammalian CTC1 and the CST complex were identified independently through analysis of the putative STN1 ortholog.28 Mass spectrometry of STN1-interacting proteins uncovered CTC1 and TEN1. Subsequent analysis demonstrated that these proteins form a trimeric complex.
Although S. pombe Stn1-Ten1 and Arabidopsis and mammalian CST clearly localize to telomeres and play a role in telomere maintenance, the level of sequence conservation between proteins identified as being homologous to Stn1, Ten1 or CTC1 is extremely low. Thus, one has to ask whether these proteins are true orthologs of S. cerevisiae Cdc13, Stn1 and Ten1. For Cdc13 and CTC1, this is still an open question as structural information is available for only a single OB-fold from Cdc13. Since Cdc13 and CTC1 are both predicted to contain multiple OB-folds, it is possible that the two proteins will turn out to resemble each other and RPA70.
An orthologous relationship between the budding yeast Stn1 and Ten1 and STN1 and TEN1 proteins in other organisms is supported by several lines of evidence. First, the crystal structure of the SpStn1-Ten1 complex has essentially the same architecture as the Stn1-Ten1 complex from the budding yeast C. tropicalis.22 Second, similar to ScStn1 and ScTen1 the OB-folds of SpStn1 and SpTen1 resemble those of Rpa2 and Rpa3 (corresponding to HsRPA32 and HsRPA14, respectively). Finally, the C-terminal domain of SpStn1, although shorter than that of ScStn1, is also predicted to contain two WH motifs. Similarly, the C-terminal domain of hSTN1 encodes at least one predicted WH domain, and NMR analysis confirms the existence of this motif in mouse STN1 (PDB1wj5). Interestingly, AtStn1 lacks a C-terminal domain altogether (Fig. 2). In ScStn1 the C-terminal domain is required for telomere length control, but plays no detectable role in telomere capping.14 Since the OB-fold domain and adjacent α-helix mediate the interaction between STN1 and TEN1,22 it is possible that the WH motif is required for a function that was lost in the 1.5 billion years since plants, humans and yeasts shared a common ancestor.
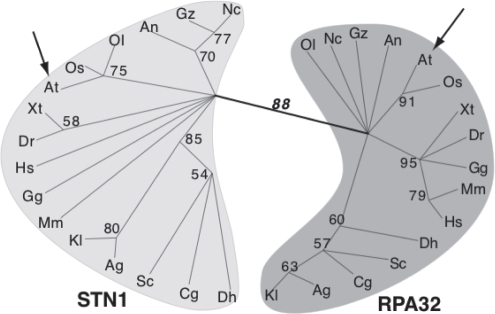
Separate domains in multi-domain proteins often have different evolutionary histories.29,30 Phylogenetic analysis of the OB-fold domains of STN1 and RPA32 (Rpa2) from 16 different eukaryotes (Fig. 3) indicates that STN1 and RPA32 form distinct monophyletic groups: ScStn1 and all other putative Stn1 orthologs, including AtSTN1, form a statistically well-supported clade while the putative RPA32 orthologs form a second separate clade. Thus, all identified STN1 proteins are likely orthologs; the ability to bind TEN1 is conserved and for all STN1 sequences identified using bioinformatic approaches, their OB-fold domains are related by common ancestry.
Figure 3.
Stn1 and Rpa32 cluster in distinct monophyletic groups. Shown is an unrooted maximum likelihood phylogeny of the OB-fold domains of STN1 and RPA32 inferred using the WAG amino-acid transition model in RAxML57 from the alignment of Gao et al.21 with the addition of STN1 from plants and green algae. Numbers along branches are bootstrap percentages from 500 replicates and indicate that STN1 and RPA32 form distinct monophyletic groups. Arrows indicate the placement of Arabidopsis. Other species are: Ag, Ashbya gossypii; An, Aspergillus nidulans; Cg, Candida glabrata; Dh, Debaryomyces hansenii; Dr, Danio rerio; Gg, Gallus gallus; Gz, Gibberella zeae; Hs, Homo sapiens; Kl, Kluyveromyces lactis; Nc, Neurospora crassa; Mm, Mus musculus; Os, Oryza sativa; Ol, Ostreococcus lucimarinus; Sc, Saccharomyces cerevisiae; Xt, Xenopus tropicalis.
From a functional standpoint, evidence is also emerging that the various activities of CST are conserved, although there is there is considerable species to species variation in the relative importance of each activity. Both S. pombe and Arabidopsis STN1 play an essential role in telomere protection,25,26 and as discussed below, Arabidopsis CST may also contribute to telomeric DNA replication. In contrast, the vertebrate CST appears to have lost its essential function in telomere protection perhaps as shelterin components emerged, but it has retained a specialized role in telomere replication.
Plant CST
CTC1 localizes to telomeres in both cycling and non-cycling Arabidopsis cells as expected for a protein involved in chromosome end protection.27 Although null mutations in telomere capping proteins are lethal events in mammalian cells and even yeast (e.g., loss of CDC13, STN1 or TEN1 in budding yeast, STN1, TEN1 or POT1 in fission yeast and TRF2 or POT1 in vertebrates1), Arabidopsis mutants lacking STN1 and CTC1 are viable at least initially.26,27 Telomere length in null mutants is much more heterogeneous, and overall significantly shorter than in wildtype plants. 20–35% of mitotic cells from mutant pistils contain end-to-end chromosome fusions, and these are predominately subtelomere-to-subtelomere fusion events. In addition, extrachromosomal telomeric circles, a marker for telomere recombination, are observed in null mutants, arguing that telomere stability is decreased in the absence of CST components.
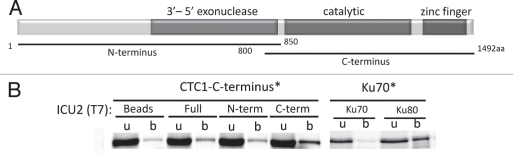
As in yeast and vertebrate CST mutants, plants lacking CTC1 or STN1 display increased G-overhang signals, in this case three to four times higher than in wild-type. While this phenotype could result from loss of C-strand protection, it is also possible that the Arabidopsis CST has an additional function in telomere replication. In vitro co-immunoprecipitation experiments support this conclusion by revealing an interaction between the C-terminus of CTC1 and the C-terminus of the catalytic subunit of Polα, Incurvata2 (ICU2)31 (Fig. 4). These data indicate that the physical association between CST and lagging strand replication machinery is conserved and further that the Arabidopsis CST complex may participate in both telomere capping and telomeric DNA replication.
Figure 4.
Arabidopsis CTC1 interacts with the catalytic subunit of DNA polymerase α (ICU2) in vitro. (A) Diagram of DNA polymerase α domain structure. (B) Co-immunopreciptiation was conducted with 35S-Met labeled (asterisk) and T7-tagged unlabeled protein expressed in rabbit reticulocyte lysate. The C-terminal half of AtCTC1 was used for binding reactions with different regions of ICU2. When bound to a tagged partner, labeled protein is precipitated on T7-beads (b) from the unbound supernatant (u) fraction. Ku70/80 interaction served as a positive control.
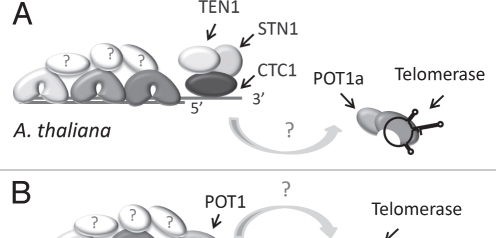
Only a subset of the vertebrate shelterin components have been identified in plants; no RAP1, TPP1 or TIN2 can be discerned. Arabidopsis encodes multiple TRF-like proteins, which bind ds telomeric DNA in vitro and negatively regulate telomere length in vivo32,33 (Fig. 5A). Notably, the two POT1 paralogs in Arabidopsis, POT1a and POT1b, do not bind single-strand telomeric DNA in vitro34 and do not function in telomere capping35 (Nelson A., Shakirov E. and Shippen D., unpublished data). Rather, these proteins associate with the telomerase RNP35,36 (Fig. 5A). These findings raise the possibility that chromosome end protection proceeds by a fundamentally different mechanism in plants versus other eukaryotes. This hypothesis is refuted by recent analysis of POT1 from the moss Physcomitrella patens.37 P. patens POT1 binds G-rich telomeric DNA in vitro and is critical for chromosome end protection in vivo. Thus, the telomere capping function of POT1 is conserved in early diverging land plants, but appears to have been lost in Arabidopsis. P. patens also encodes STN1, TEN1 and CTC1 orthologs (Fig. 5B). The extent to which these CST components contribute to chromosome end protection, telomeric DNA replication or both processes remains to be determined. Nevertheless, the current data suggest that plants represent an evolutionary bridge between S. cerevisiae, which has only CST as a capping complex and vertebrates, which use shelterin for end protection and CST for special replication functions.
Figure 5.
Model for telomere capping complexes in the flowering plant Arabidopsis thaliana and the moss Physcomitrella patens. (A) CST functions as the major telomere capping complex in Arabidopsis. Multiple TRF-like proteins have been described, but other shelterin-like components cannot be identified in plant genomes. POT1a is a telomerase accessory factor and is not required for chromosome end protection. (B) P. patens encodes two TRF-like proteins and a single POT1 protein. The moss POT1 binds single-stranded G-rich telomeric DNA and functions in a manner analogous to vertebrate and yeast POT1. CST components are encoded in the P. patens genome, but their function is unknown.
Mammalian CST
While mouse and human CST localize to telomeres as shown by both indirect immunofluorescence and chromatin immunoprecipitation, several lines of evidence suggest that mammalian CST has both telomeric and non-telomeric functions.28 First, only ∼20% of CST-containing foci localize to telomeres. The remaining ∼80% are present in the nucleus but the identity of these foci is unclear. They do not correspond to replication foci and the relative distribution of CST between telomeric and non-telomeric locations is unaffected by cell cycle stage.
The DNA binding properties of mammalian CST also suggest non-telomeric as well as telomeric functions.28 The complex binds specifically to ssDNA, but the interaction is not sequence specific and all three subunits are required for high affinity binding (nM Kd). Moreover, the minimum binding site size is large and binding affinity increases with increasing length of DNA (20 nt<<34 nt<74 nt). This contrasts with the situation in budding yeast where binding of CST is sequence-specific and only Cdc13 is required for high affinity binding to the well defined 11 nt consensus sequence.17 The DNA-binding properties of mammalian CST more closely resemble those of RPA, which also lacks sequence specificity and prefers an extended DNA binding site.23,38 The extended RPA binding site (28–32 nt) is thought to reflect the sequential binding of multiple OB-folds along the length of the DNA. It will be interesting to learn whether this also occurs with CST.
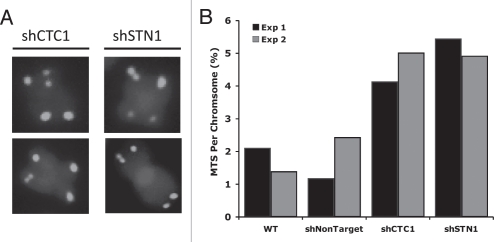
Depletion of either CTC1 or STN1 results in a variety of telomeric and nontelomeric defects.27,28 RNAi depletion of either protein causes an elevation in H2AX phosphorylation and an increase in anaphase bridges and chromatin bridges between newly separated daughter cells. The γH2AX foci do not localize to telomeres and telomere fusions between metaphase chromosomes are rarely observed. Thus, CTC1 or STN1 knockdown seems to cause a general increase in genome instability rather than a loss of telomere protection. Other knockdown phenotypes indicate a role for CST in telomere replication. Telomere FISH with cells depleted of either CTC1 or STN1 shows an increase in telomeres with aberrant structure and telomeres with multiple hybridization signals are particularly apparent (Fig. 6). Such multi-telomeric signals indicate discontinuities in the telomeric chromatin (akin to fragile sites) and are a hallmark of defects in telomere replication.39 Thus, the appearance of multi-telomeric signals after CTC1 or STN1 knockdown suggests mammalian CST may be important for replication of the duplex region of the telomeric tract. CTC1 or STN1 knockdown also causes an increase in single-stranded (ss) telomeric DNA.27,28 Analysis by non-denaturing hybridization reveals that a portion of this ssDNA is resistant to exonuclease 1 digestion and hence must occur within the duplex region of the telomere. Accumulation of ssDNA within the telomeric tract fits with replication defects that lead to replication fork stalling. The remainder of the ssDNA corresponds to an increase in G-overhang length. This phenotype again suggests a defect associated with telomere replication such as increased G-overhang processing, increased telomerase action or decreased C-strand fill-in.40
Figure 6.
Knockdown of human CST components, CTC1 or STN1, results in multi-telomeric signals (MTS). (A) Examples of MTS in stable shRNA knockdown clones of either CTC1 or STN1. Telomeric PNA-FITC probe (green); DAPI (blue). (B) Quantification of MTS. Black and gray bars represent independent experiments.
Although CST does not appear to play a primary role in telomere protection, it can complement the protective function of POT1/TPP1.28 Knockdown of POT1 causes an increase in the number of telomeres with DNA damage signals (TIFs) and this is exacerbated if both POT1 and STN1 are depleted even though no TIFs are observed after depletion of STN1 alone. Likewise, a POT1/STN1 double knockdown causes an even greater increase in G-overhang length than the single STN1 knockdown.
Independent evidence that CST contributes to telomere maintenance in humans comes from a large genomewide association study looking for SNPs associated with fluctuation in leukocyte telomere length.41 In this study involving 3,417 participants, only two new genes were identified as having association at a genome-wide significance level. One of these was STN1 (otherwise known as OBFC1). Evidence for association was also obtained for regions of TERC, the telomerase RNA subunit. The association between STN1/OBFC1 and leukocyte telomere length was replicated using de novo genotyping and a retrospective look-up analysis of data from additional individuals.
Mechanism of Action
Clues concerning the mechanism of CST action in mammalian cells come from analysis of a DNA Polα accessory factor called AAF (alpha accessory factor) that was recently found to be composed of CTC1 and STN1.27,28,42 The complex of AAF132/CTC1 and AAF44/STN1 was originally identified as a factor that copurified with DNA Polα.43 Subsequent biochemical studies indicated that AAF/CTC1-STN1 functions by increasing the affinity of DNA Polα/primase for template DNA, leading to the suggestion that AAF/CTC1-STN1 might assist DNA Polα/primase in synthesis of the lagging strand of a replication fork.44 Given the well established role of S. cerevisiae Cdc13 and Stn1 in recruiting DNA Polα/primase to fill in the complementary C-strand following telomerase action,7 it is striking that both mammalian and Arabidopsis CTC1 and/or STN1 associate with DNA Polα/primase (Fig. 4; reviewed in ref. 27) and the mammalian proteins act as a template affinity factor.44 These findings lead us to suggest that plant and mammalian CST may also serve to recruit DNA Polα/primase for C-strand fill-in (Fig. 7). The need for a specialized mechanism to recruit DNA-Polα to the chromosome terminus is anticipated because telomerase acts after the replication fork reaches the chromosome terminus.45,46 Hence, the components of the replication progression complex that normally recruit DNA Polα/primase to the replication fork (And1 and Mcm10)47–49 are unlikely to remain at the chromosome terminus at the time of C-strand fill in.46
Figure 7.
Model for CST in telomeric DNA replication in budding yeast and vertebrates. S. cerevisiae CST interacts with the Est1 component of telomerase to promote telomeric DNA synthesis on the G-overhang, and with Polα/primase to facilitate lagging strand replication of the C-strand. Vertebrate CST associates with Polα/primase and stimulates its priming activity. The shelterin component TPP1 contacts telomerase and is postulated to recruit it to the chromosome terminus. TPP1 may also recruit CST to the telomere via interactions with STN1.
One obvious question concerns how CST is loaded at the telomere given its lack of DNA-binding specificity. A recent study indicates that human STN1 interacts with TPP1,50 suggesting that in mammalian cells CST loading may occur through interactions with shelterin (Fig. 7). Given that the telomeric function of mammalian CST was discovered only recently, it is also possible that CST is brought to the chromosome terminus through interactions with as yet unidentified telomere proteins. TPP1 also interacts with telomerase51,52 and enhances its action by increasing enzyme processivity.53 Thus, TPP1 could potentially function in a manner similar to Cdc13 by serving to first enhance telomerase activity and then to recruit DNA Polα/primase via interactions with CST.
The current data argue that mammalian CST may have both telomeric and non-telomeric functions. Specifically, this complex may serve as a DNA Polα/primase recruitment factor elsewhere in the genome (Fig. 8). Conditions that promote replication stress, for example replication through highly repetitive sequences or after certain types of DNA damage, will cause the polymerase to become uncoupled from the MCM1–7 replicative helicase. This scenario would lead to accumulation of ssDNA and a need to re-initiate leading or lagging strand synthesis. Recruitment of CST to stalled replication forks could stimulate synthesis by Polα/primase to restart replication and thus contribute to global genome stability.
Figure 8.
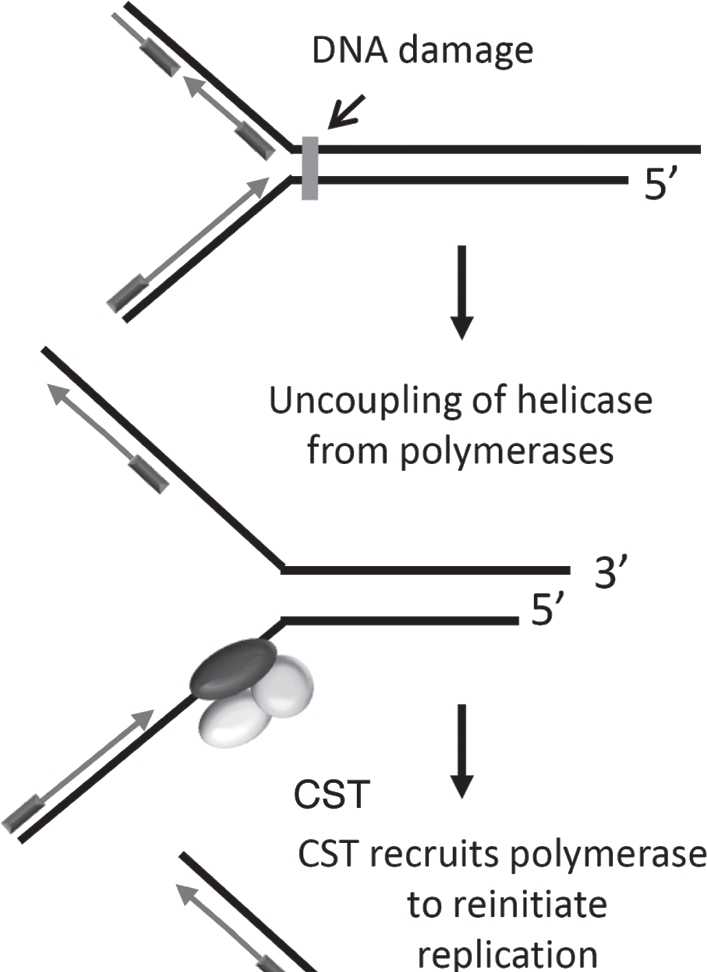
Model for CST function during replication of non-telomeric DNA. Replication stress (following DNA damage or synthesis through highly repetitive sequences) results in polymerase dissociation from replicative helicases. CST may recruit and stimulate the activity of DNA polα/primase to promote lagging strand replication at such sites.
Evolution of CST Function
It is striking how CST complexes from different organisms perform similar activities in telomere protection and aspects of DNA replication, but in any one organism only a subset of these activities predominate. In terms of telomere protection, budding yeast and mammalian cells represent the two ends of the spectrum as CST is essential in S. cerevisiae, while in human cells it only contributes when POT1/TPP1 is depleted. Plants provide a fascinating middle ground as some have evolved to use CST as their main telomere protection complex, while others have retained the capping function of POT1 (and likely other shelterin components). The current data indicate that budding yeast CST function is confined to replication of the extreme chromosome terminus. However, it is conceivable that ScCST has additional replication/repair functions resembling those of the mammalian complex. Such functions may not have been uncovered because removal of CST subunits leads to such a severe and immediate telomere uncapping phenotype. In support of this idea, when overexpressed, STN1 localizes to replication forks and interferes with the S-phase checkpoint in a DNA Polα-dependant manner.54
Finally, the structural similarity between CST and RPA is remarkable given that the two protein complexes have distinct biological roles. The similarity suggests that CST may resemble RPA in having multiple interaction partners and alternative DNA binding modes that involve a variable number of OB-folds. Such diversity in interactions might allow CST to mediate the sequential events that take place during telomere replication in much the same way that RPA mediates nucleic acid transactions during DNA replication, recombination and repair.23,55 It is noteworthy that an RPA-like protein Teb1 is a key component of the telomerase holoenzyme from Tetrahymena.56 The OB-folds from Teb1 bind telomeric G-strand DNA, thus anchoring telomerase and allowing enzyme processivity. Again the sequential binding of the multiple OB-folds may be important as this could prevent Pot1 binding as the telomere is extended. Thus, RPA-like proteins may be much more common than originally thought and play key roles in a wide variety of chromosomal processes.
Acknowledgements
Work from our labs is supported by NIH grants GM065383 (D.E.S.), GM041803 (C.M.P.), GM088728 (C.M.P), NSF grant MCB-0843399 (D.E.S.) and Ruth L. Kirschstein National Research Service Awards F32 CA117846 (J.A.S.) and F32 GM093635 (M.A.B.).
Footnotes
Previously published online: www.landesbioscience.com/journals/cc/article/12547
References
- 1.Palm W, de Lange T. How shelterin protects mammalian telomeres. Annu Rev Genet. 2008;42:18–20. doi: 10.1146/annurev.genet.41.110306.130350. [DOI] [PubMed] [Google Scholar]
- 2.O'Sullivan RJ, Karlseder J. Telomeres: Protecting chromosomes against genome instability. Nat Rev Mol Cell Biol. 2010;11:171–181. doi: 10.1038/nrm2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lei M, Podell ER, Cech TR. Structure of human POT1 bound to telomeric single-stranded DNA provides a model for chromosome end-protection. Nat Struct Mol Biol. 2004;11:1223–1229. doi: 10.1038/nsmb867. [DOI] [PubMed] [Google Scholar]
- 4.Linger BR, Price CM. Conservation of telomere protein complexes: Shuffling through evolution. Crit Rev Biochem Mol Biol. 2009;44:434–446. doi: 10.3109/10409230903307329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miyoshi T, Kanoh J, Saito M, Ishikawa F. Fission yeast Pot1-Tpp1 protects telomeres and regulates telomere length. Science. 2008;320:1341–1344. doi: 10.1126/science.1154819. [DOI] [PubMed] [Google Scholar]
- 6.Lundblad V. Budding yeast telomeres. In: de Lange T, Lundblad V, Blackburn E, editors. Telomeres. Cold Spring Harbor, NY: Cold Spring Harbor Press; 2006. pp. 345–386. [Google Scholar]
- 7.Bianchi A, Shore D. How telomerase reaches its end: Mechanism of telomerase regulation by the telomeric complex. Mol Cell. 2008;31:153–165. doi: 10.1016/j.molcel.2008.06.013. [DOI] [PubMed] [Google Scholar]
- 8.Garvik B, Carson M, Hartwell L. Single-stranded DNA arising at telomeres in cdc13 mutants may constitute a specific signal for the RAD9 checkpoint. Mol Cell Biol. 1995;15:6128–6138. doi: 10.1128/mcb.15.11.6128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Grandin N, Damon C, Charbonneau M. Ten1 functions in telomere end protection and length regulation in association with Stn1 and Cdc13. EMBO J. 2001;20:1173–1183. doi: 10.1093/emboj/20.5.1173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grandin N, Reed SI, Charbonneau M. Stn1, a new Saccharomyces cerevisiae protein, is implicated in telomere size regulation in association with Cdc13. Genes Dev. 1997;11:512–527. doi: 10.1101/gad.11.4.512. [DOI] [PubMed] [Google Scholar]
- 11.Li S, Makovets S, Matsuguchi T, Blethrow JD, Shokat KM, Blackburn EH. Cdk1-dependent phosphorylation of Cdc13 coordinates telomere elongation during cell cycle progression. Cell. 2009;136:50–61. doi: 10.1016/j.cell.2008.11.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chan A, Boule JB, Zakian VA. Two pathways recruit telomerase to Saccharomyces cerevisiae telomeres. PLoS Genet. 2008;4:1000236. doi: 10.1371/journal.pgen.1000236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.DeZwaan DC, Freeman BC. The conserved Est1 protein stimulates telomerase DNA extension activity. Proc Natl Acad Sci USA. 2009;106:17337–17342. doi: 10.1073/pnas.0905703106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Puglisi A, Bianchi A, Lemmens L, Damay P, Shore D. Distinct roles for yeast Stn1 in telomere capping and telomerase inhibition. EMBO J. 2008;27:2328–2339. doi: 10.1038/emboj.2008.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grossi S, Puglisi A, Dmitriev PV, Lopes M, Shore D. Pol12, the B subunit of DNA polymerase alpha, functions in both telomere capping and length regulation. Genes Dev. 2004;18:992–1006. doi: 10.1101/gad.300004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Qi H, Zakian VA. The Saccharomyces telomerebinding protein Cdc13p interacts with both the catalytic subunit of DNA polymerase alpha and the telomerase-associated est1 protein. Genes Dev. 2000;14:1777–1788. [PMC free article] [PubMed] [Google Scholar]
- 17.Mitton-Fry RM, Anderson EM, Hughes TR, Lundblad V, Wuttke DS. Conserved structure for single-stranded telomeric DNA recognition. Science. 2002;296:145–147. doi: 10.1126/science.1068799. [DOI] [PubMed] [Google Scholar]
- 18.Churikov D, Wei C, Price CM. Vertebrate POT1 restricts G-overhang length and prevents activation of a telomeric DNA damage checkpoint but is dispensable for overhang protection. Mol Cell Biol. 2006;26:6971–6982. doi: 10.1128/MCB.01011-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hockemeyer D, Daniels JP, Takai H, de Lange T. Recent expansion of the telomeric complex in rodents: Two distinct POT1 proteins protect mouse telomeres. Cell. 2006;126:63–77. doi: 10.1016/j.cell.2006.04.044. [DOI] [PubMed] [Google Scholar]
- 20.Kibe T, Osawa GA, Keegan CE, de Lange T. Telomere protection by TPP1 is mediated by POT1a and POT1b. Mol Cell Biol. 2010;30:1059–1066. doi: 10.1128/MCB.01498-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao H, Cervantes RB, Mandell EK, Otero JH, Lundblad V. RPA-like proteins mediate yeast telomere function. Nat Struct Mol Biol. 2007;14:208–214. doi: 10.1038/nsmb1205. [DOI] [PubMed] [Google Scholar]
- 22.Sun J, Yu EY, Yang Y, Confer LA, Sun SH, Wan K, et al. Stn1-Ten1 is an Rpa2-Rpa3-like complex at telomeres. Genes Dev. 2009;23:2900–2914. doi: 10.1101/gad.1851909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fanning E, Klimovich V, Nager AR. A dynamic model for replication protein A (RPA) function in DNA processing pathways. Nucleic Acids Res. 2006;34:4126–4137. doi: 10.1093/nar/gkl550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gelinas AD, Paschini M, Reyes FE, Heroux A, Batey RT, Lundblad V, et al. Telomere capping proteins are structurally related to RPA with an additional telomere-specific domain. Proc Natl Acad Sci USA. 2009;106:19298–19303. doi: 10.1073/pnas.0909203106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Martin V, Du LL, Rozenzhak S, Russell P. Protection of telomeres by a conserved Stn1-Ten1 complex. Proc Natl Acad Sci USA. 2007;104:14038–14043. doi: 10.1073/pnas.0705497104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Song X, Leehy K, Warrington RT, Lamb JC, Surovtseva YV, Shippen DE. STN1 protects chromosome ends in Arabidopsis thaliana. Proc Natl Acad Sci USA. 2008;105:19815–19820. doi: 10.1073/pnas.0807867105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Surovtseva YV, Churikov D, Boltz KA, Song X, Lamb JC, Warrington R, et al. Conserved telomere maintenance component 1 interacts with STN1 and maintains chromosome ends in higher eukaryotes. Mol Cell. 2009;36:207–218. doi: 10.1016/j.molcel.2009.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miyake Y, Nakamura M, Nabetani A, Shimamura S, Tamura M, Yonehara S, et al. RPA-like mammalian Ctc1-Stn1-Ten1 complex binds to single-stranded DNA and protects telomeres independently of the Pot1 pathway. Mol Cell. 2009;36:193–206. doi: 10.1016/j.molcel.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 29.Richards TA, Cavalier-Smith T. Myosin domain evolution and the primary divergence of eukaryotes. Nature. 2005;436:1113–1118. doi: 10.1038/nature03949. [DOI] [PubMed] [Google Scholar]
- 30.Koonin EV, Aravind L, Kondrashov AS. The impact of comparative genomics on our understanding of evolution. Cell. 2000;101:573–576. doi: 10.1016/s0092-8674(00)80867-3. [DOI] [PubMed] [Google Scholar]
- 31.Barrero JM, Gonzalez-Bayon R, delPozo JC, Ponce MR, Micol JL. ICURVATA2 encodes the catalytic subunit of DNA polymerase alpha and interacts with genes involved in chromatin-mediated cellular memory in Arabidopsis thaliana. Plant Cell. 2007;19:2822–2838. doi: 10.1105/tpc.107.054130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hong JP, Byun MY, Koo DH, An K, Bang JW, Chung IK, et al. Suppression of Rice Telomere Binding Protein 1 results in severe and gradual developmental defects accompanied by genome instability in rice. Plant Cell. 2007;19:1770–1781. doi: 10.1105/tpc.107.051953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Karamysheva ZN, Surovtseva YV, Vespa L, Shakirov EV, Shippen DE. A C-terminal Myb extension domain defines a novel family of double-strand telomeric DNA-binding proteins in Arabidopsis. J Biol Chem. 2004;279:47799–47807. doi: 10.1074/jbc.M407938200. [DOI] [PubMed] [Google Scholar]
- 34.Shakirov EV, McKnight TD, Shippen DE. POT1-independent single-strand telomeric DNA binding activities in Brassicaceae. Plant J. 2009;58:1004–1015. doi: 10.1111/j.1365-313X.2009.03837.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Surovtseva YV, Shakirov EV, Vespa L, Osbun N, Song X, Shippen DE. Arabidopsis POT1 associates with the telomerase RNP and is required for telomere maintenance. EMBO J. 2007;26:3653–3661. doi: 10.1038/sj.emboj.7601792. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cifuentes-Rojas C, Kannan K, Tseng L, Shippen DE. Arabidopsis TER1 assembles with Pot1a for telomere maintenance. In preparation. [Google Scholar]
- 37.Shakirov EV, Perroud P-F, Nelson AD, Cannell ME, Quatrano RS, Shippen DE. Protection of telomeres 1 is required for telomere integrity in the moss Physcomitrella patens. Plant Cell. 2010;22:1838–1848. doi: 10.1105/tpc.110.075846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wold MS. Replication protein A: A heterotrimeric, single-stranded DNA-binding protein required for eukaryotic DNA metabolism. Annu Rev Biochem. 1997;66:61–92. doi: 10.1146/annurev.biochem.66.1.61. [DOI] [PubMed] [Google Scholar]
- 39.Sfeir A, Kosiyatrakul ST, Hockemeyer D, MacRae SL, Karlseder J, Schildkraut CL, et al. Mammalian telomeres resemble fragile sites and require TRF1 for efficient replication. Cell. 2009;138:90–103. doi: 10.1016/j.cell.2009.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Verdun RE, Karlseder J. Replication and protection of telomeres. Nature. 2007;447:924–931. doi: 10.1038/nature05976. [DOI] [PubMed] [Google Scholar]
- 41.Levy D, Neuhausen SL, Hunt SC, Kimura M, Hwang SJ, Chen W, et al. Genome-wide association identifies OBFC1 as a locus involved in human leukocyte telomere biology. Proc Natl Acad Sci USA. 2010;107:9293–9298. doi: 10.1073/pnas.0911494107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Casteel DE, Zhuang S, Zeng Y, Perrino FW, Boss GR, Goulian M, et al. A DNA polymerase-alpha/primase cofactor with homology to replication protein A-32 regulates DNA replication in mammalian cells. J Biol Chem. 2009;284:5807–5818. doi: 10.1074/jbc.M807593200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Goulian M, Heard CJ, Grimm SL. Purification and properties of an accessory protein for DNA polymerase alpha/primase. J Biol Chem. 1990;265:13221–13230. [PubMed] [Google Scholar]
- 44.Goulian M, Heard CJ. The mechanism of action of an accessory protein for DNA polymerase alpha/primase. J Biol Chem. 1990;265:13231–13239. [PubMed] [Google Scholar]
- 45.Moser BA, Subramanian L, Chang YT, Noguchi C, Noguchi E, Nakamura TM. Differential arrival of leading and lagging strand DNA polymerases at fission yeast telomeres. EMBO J. 2009;28:810–820. doi: 10.1038/emboj.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhao Y, Sfeir AJ, Zou Y, Buseman CM, Chow TT, Shay JW, et al. Telomere extension occurs at most chromosome ends and is uncoupled from fill-in in human cancer cells. Cell. 2009;138:463–475. doi: 10.1016/j.cell.2009.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chattopadhyay S, Bielinsky AK. Human Mcm10 regulates the catalytic subunit of DNA polymerase-alpha and prevents DNA damage during replication. Mol Biol Cell. 2007;18:4085–4095. doi: 10.1091/mbc.E06-12-1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gambus A, van Deursen F, Polychronopoulos D, Foltman M, Jones RC, Edmondson RD, et al. A key role for Ctf4 in coupling the MCM2-7 helicase to DNA polymerase alpha within the eukaryotic replisome. EMBO J. 2009;28:2992–3004. doi: 10.1038/emboj.2009.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhu W, Ukomadu C, Jha S, Senga T, Dhar SK, Wohlschlegel JA, et al. Mcm10 and And-1/CTF4 recruit DNA polymerase alpha to chromatin for initiation of DNA replication. Genes Dev. 2007;21:2288–2299. doi: 10.1101/gad.1585607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wan M, Qin J, Songyang Z, Liu D. OB fold-containing protein 1 (OBFC1), a human homolog of yeast Stn1, associates with TPP1 and is implicated in telomere length regulation. J Biol Chem. 2009;284:26725–26731. doi: 10.1074/jbc.M109.021105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xin H, Liu D, Wan M, Safari A, Kim H, Sun W, et al. TPP1 is a homologue of ciliate TEBP-beta and interacts with POT1 to recruit telomerase. Nature. 2007;445:559–562. doi: 10.1038/nature05469. [DOI] [PubMed] [Google Scholar]
- 52.Zaug AJ, Podell ER, Nandakumar J, Cech TR. Functional interaction between telomere protein TPP1 and telomerase. Genes Dev. 24:613–622. doi: 10.1101/gad.1881810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Latrick CM, Cech TR. POT1-TPP1 enhances telomerase processivity by slowing primer dissociation and aiding translocation. EMBO J. 2010;29:924–933. doi: 10.1038/emboj.2009.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gasparyan HJ, Xu L, Petreaca RC, Rex AE, Small VY, Bhogal NS, et al. Yeast telomere capping protein Stn1 overrides DNA replication control through the S phase checkpoint. Proc Natl Acad Sci USA. 2009;106:2206–2211. doi: 10.1073/pnas.0812605106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Haring SJ, Mason AC, Binz SK, Wold MS. Cellular functions of human RPA1. Multiple roles of domains in replication, repair and checkpoints. J Biol Chem. 2008;283:19095–19111. doi: 10.1074/jbc.M800881200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Min B, Collins K. Multiple mechanisms for elongation processivity within the reconstituted Tetrahymena telomerase holoenzyme. J Biol Chem. 2010;107:9293–9298. doi: 10.1074/jbc.M110.119172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Stamatakis A. RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics. 2006;22:2688–2690. doi: 10.1093/bioinformatics/btl446. [DOI] [PubMed] [Google Scholar]