Abstract
Many animals show intimate interactions with bacterial symbionts that provision hosts with limiting nutrients. The best studied such association is that between aphids and Buchnera aphidicola, which produces essential amino acids that are rare in the phloem sap diet. Genomic studies of Buchnera have provided a new means for inferring metabolic capabilities of the symbionts and their likely contributions to hosts. Despite evolutionary reduction of genome size, involving loss of most ancestral genes, Buchnera retains capabilities for biosynthesis of all essential amino acids. In contrast, most genes duplicating amino acid biosynthetic capabilities of hosts have been eliminated. In Buchnera of many aphids, genes for biosynthesis of leucine and tryptophan have been transferred from the chromosome to distinctive plasmids, a feature interpreted as a mechanism for overproducing these amino acids through gene amplification. However, the extent of plasmid-associated amplification varies between and within species, and plasmid-borne genes are sometimes fewer in number than single copy genes on the (polyploid) main chromosome. This supports the broader interpretation of the plasmid location as a means of achieving regulatory control of gene copy number and/or transcription. Buchnera genomes have eliminated most regulatory sequences, raising the question of the extent to which gene expression is moderated in response to changing demands imposed by host nutrition or other factors. Microarray analyses of the Buchnera transcriptome reveal only slight changes in expression of nutritionrelated genes in response to shifts in host diet, with responses less dramatic than those observed for the related nonsymbiotic species, Escherichia coli.
Symbioses between bacteria and eukaryotes, i.e., chronic infections that are part of the normal life history of the host and are often beneficial, are ubiquitous in nature but have historically received little attention from experimental biologists or ecologists. This situation has been reversed in the last few years, which have seen a surge of interest and progress in this field (1). One useful view of these symbioses is as persistent intimate associations in which partners interact through the transfer of molecules, particularly small molecules that are essential to the growth requirements of each organism. However, the interdependence of partners that makes symbiosis so intriguing also has made it difficult to study. In particular, the inability to culture most symbionts has thwarted characterization of their chemical interactions with hosts. Recently, however, the acquisition of DNA sequences, including whole genomic sequences, of symbionts has enabled major progress in defining the biosynthetic capabilities that underlie contributions to hosts.
Invertebrates generally, and insects especially, show a particularly striking variety of symbioses with bacteria. Many of these associations are conspicuous because of the presence of a “bacteriome” (or “mycetome”), a specialized structure in the host body that houses the symbionts and that can occupy a substantial proportion of the host biomass. The symbionts are often intracellular but frequently enclosed within the cytoplasm by a host-derived membrane. Bacteriome-associated symbioses of animals, with emphasis on arthropods, were studied extensively by light microscopy during the first half of the 20th century and summarized in a book by the prominent symbiosis researcher Paul Buchner (2). The volume describes a kind of fairyland of intimate and highly specialized biological interactions, many of which have not been further studied. Despite the anatomical and histological diversity of these symbiotic arrangements, Buchner proposed a unified functional role for bacteriome occupants, hypothesizing that their raison d'etre is the provisioning of nutrients that animals are unable to synthesize themselves and that are absent or limiting in the specialized diets exploited by particular animal groups. Extensive sections of his book are devoted to insects with highly restricted diets, such as plant sap or vertebrate blood. Although nutritional provisioning has been better documented for some associations than others, the overall evidence supports the view that symbiosis has often enabled the expansion of animal niches and has contributed substantially to evolutionary diversification. The macroevolutionary role of nutrition-based symbiosis can be appreciated by recognizing that insects feeding on phloem or xylem sap, including aphids, psyllids, whiteflies, scale insects, planthoppers, cicadas, spittlebugs, and most leafhoppers, are dependent for their way of life on obligate bacterial endosymbionts.
Recently, molecular approaches have enabled substantial progress in understanding the evolution and functioning of the obligate symbionts of several insect hosts. Based on DNA sequence data, we now know that intracellular, bacteriome-associated symbiotic associations of insects, involving vertical transmission through eggs, typically descend from ancient infections of ancestors dating back 100 million years or more (1). Phylogenetic studies have also revealed that many of the intracellular symbionts within insects are closely related to the well studied experimental organism, Escherichia coli, for which functional information is available for many gene products. This fact has provided an ideal basis for using gene sequences to infer symbiont metabolic capabilities, including their contributions to host nutrition.
Here we consider the question of how a bacterial genome has been modified in the context of the new demands of symbiosis, especially nutrient provisioning to host tissues. We focus on the best-studied insect symbiont, Buchnera aphidicola, which has coevolved with its aphid hosts for >100 million years (3). We have divided the question into two main aspects: (i) modifications of genome contents and organization and (ii) modification of controls on gene expression. Because different hosts encounter varying nutritional demands and varying dietary conditions, we expect some heritable modifications manifested as differences among species or populations. In addition, individual host species must deal with a variety of nutritional limitations during the course of a season or while using different resources or habitats; thus, we might expect that symbiont productivity can be modified as a result of environmental factors.
Nutritional Provisioning as a Primary Task of Symbionts in Sap-Feeders
Animals generally are lacking in pathways underlying amino acid production, typically requiring 10 “essential” amino acids in the diet; this limitation must stem from ancient gene loss in an ancestor of Metazoa. Animals also have a large number of dietary requirements for enzymatic cofactors (or “vitamins”). Studies of phloem sap composition, based on collections from the severed mouthparts of aphids, show that nitrogen is almost exclusively present as amino acids, and that nonessential amino acids predominate (4). In general, the essential amino acids are present in relative concentrations considerably lower than those found in aphid proteins or in dietary requirements determined for insects. This imbalance implies that insect growth potential can be enhanced by symbionts, which absorb abundant amino acids and sugars from the host and use these to generate limiting amino acids through biosynthetic capabilities lacking in insects. However, both the total concentrations of pooled amino acids as well as the relative concentrations of individual essential amino acids can differ dramatically among plant species and phenological stages (e.g., refs. 5 and 6). Amino acid levels and profile also fluctuate, depending on the plant response to aphid feeding (7). Experimental studies have provided some evidence that the nonessential amino acid glutamate is the major nitrogenous compound exported from the aphid and used by Buchnera for production of the essential, limiting amino acids (8–10).
Reading the Genome Sequences of Insect Symbionts: Old Tools for New Tasks
Complete genome sequences are available for Buchnera of three distantly related aphid species [Acyrthosiphon pisum (11), Schizaphis graminum (12), and Baizongia pistaciae (13)], for Wigglesworthia brevipalpis, the obligate symbiont of tsetse fly (14), and for Blochmannia camponotii, the obligate symbiont of carpenter ants (15). The genome sequences of several other insect symbionts are expected to be available soon. Genomic sequences allow us to identify which pathways symbionts possess, and, even more uniquely, which pathways they lack. This information has greatly extended our knowledge of the nutritional role of symbionts within hosts and provides a foundation for further studies of chemical exchange between host and symbiont.
Based on the systems studied so far, bacteriome associates in insects show extensive genomic modifications, including massive loss of ancestral genes and many rearrangements affecting order and orientation of genes on the chromosome. For Buchnera, Wigglesworthia, and Baumannia, genomes are reduced to only ≈20% of the genes present in ancestors and modern relatives, and each contains many unique gene arrangements, yielding only small regions of synteny with related bacteria (15–17). Furthermore, in contrast to most other bacterial genomes including those of its closest relatives, these symbiont genomes show little evidence of gene uptake. Indeed, Buchnera lineages appear to lack any novel genes not present in related free-living bacteria, and comparisons of Buchnera genomes, that diverged with their ancestors >100 million years ago, show lower levels of gene acquisition or genomic rearrangements than do any other fully sequenced bacteria (11).
The combination of genome reduction and absence of gene acquisition implies that, in these insect symbioses, the basis for the symbiotic contributions lies in ancestral genes and pathways, present in free-living relatives, that were coopted for the purpose of provisioning hosts after the initiation of symbiosis. For both Buchnera and Wigglesworthia, the maintenance of ancestral genes for nutrient biosynthesis provides a clear indication of their mutualistic relationship to hosts and sharply contrasts with the loss of these same pathways from genomes of obligately parasitic bacteria, which acquire metabolites from host tissues (1). Buchnera of A. pisum has genes for all of the essential amino acid pathways (total of 54 loci), plus genes for fixation of inorganic sulfur and for synthesis of riboflavin (13). Because unneeded genes are quickly eliminated from these genomes, the retention of these biosynthetic pathways gives a clear indication of a continued contribution of the corresponding nutrients to hosts.
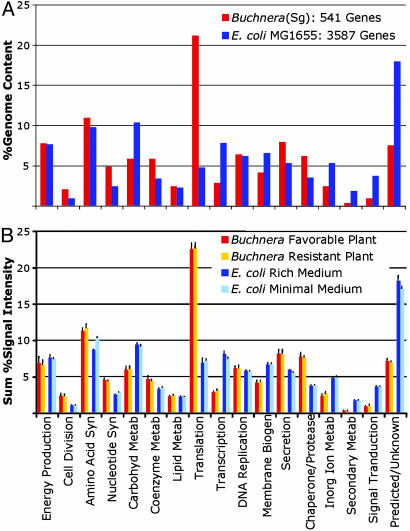
The prominence of essential amino acid production as an activity of Buchnera is evident from the set of genes retained (Fig. 1A). The E. coli genome encodes ≈60 genes for the core biosynthetic pathways underlying production of essential amino acids, ≈20 for the core pathways for nonessential amino acids plus >50 genes involved in transport of amino acids. Of these, Buchnera retains almost all of the genes for production of essential amino acids and only two (or four, depending on how amino acids are categorized) for synthesis of nonessential amino acids or amino acid transport (13). Overall, the core genes for biosynthesis of essential amino acids comprise a larger proportion (≈10%) of the Buchnera genome than of the E. coli genome (<2%).
Fig. 1.
(A) Proportional representation of genes grouped by functional categories within the genomes of Buchnera (S. graminum) and E. coli, based on functional categorizations established for the Clusters of Orthologous Groups (COGs) (52). (B) Representation of COG functional categories in the transcriptomes of Buchnera (S. graminum) and E. coli. Buchnera were from mixed-age host aphids grown at 20°C on plants differing in phloem concentrations of total and essential amino acids (barley versus resistant wheat cultivar; plants as in ref. 7). E. coli data are from the public ASAP database (53) and depict transcriptomes of early log-phase cultures grown in amino acid “Rich” LB medium versus “Minimal” Mops medium plus 0.2% glucose (J. Glasner, personal communication). Microarray attributes, RNA extraction procedures, and data normalization steps were similar for both bacterial species (42, 49). Bar height is the sum of gene signals in a given COG divided by the number of genes in that COG; bars are standard deviations based on experimental replicates (five replicates per Buchnera treatment; two and six replicates for E. coli LB and Minimal Media treatments).
How Are Genomes of Obligate Symbionts That Provision Hosts Modified Relative to Those of Free-Living Relatives?
The large majority of genes underlying amino acid biosynthetic pathways are confined to the main Buchnera chromosome. However, one of the early striking discoveries about the Buchnera genome, seemingly linked to the nutrient-provisioning role, was the finding that rate-limiting genes for biosynthesis of tryptophan (trpEG, ref. 18) and the genes for biosynthesis of leucine (leuABCD, ref. 19) are encoded on two distinct types of multicopy plasmids. Each of these plasmids has apparently evolved only one or very few times (20–22), and some earlybranching Buchnera lineages retain these genes on the main chromosome in an ancestral position (e.g., refs. 12 and 23).
The initial interpretation of the plasmid position of trpEG and leuABCD was that it enabled overproduction of the end products for host nutrition, because the plasmid location allowed amplification of gene copy number relative to single copy chromosomal genes. The subsequent discovery that each Buchnera cell contains multiple chromosomal copies (24) raised the more general interpretation of the plasmid location of trpEG and leuABCD as an arrangement allowing either increase or decrease in copy number relative to chromosomal genes (25), and perhaps allowing regulation of the level of amplification in response to environmental conditions. Ratios of plasmid-borne trpEG and leuABCD copies to chromosomal gene copies do vary among species, possibly in a direction that is related to host needs (26). Table 1 summarizes previously published and newly estimated ratios of plasmid-borne amino acid biosynthetic genes to single copy chromosomal genes in Buchnera of different aphid strains and species. The ratios of plasmid-borne genes to single-copy chromosomal genes range from much less than 1-fold (implying fewer plasmid copies than chromosome copies within a cell) to >10-fold, with rather similar ranges for relative amplification of both leu and trp genes (ref. 26 and Table 1). Values vary both within and between species, with at least part of the variation heritable (e.g., ref. 27). Thus, the ratios of copies of trpE to trpB (a single copy chromosomal gene) appear stable among different aphids of the same maternal line (27). Also, the amplification level may be quite constant for some species and more variable for others, based on analyses of geographically dispersed isolates of several species (refs. 25, 27, and 28 and Table 1).
Table 1. The ratios of copies of plasmid-borne amino acid biosynthetic genes (leuABCD, trpEG) to chromosomal gene copies for Buchnera of different aphid species and strains.
| Aphid host | Aphid clone/population | leuABCD | trpEG |
|---|---|---|---|
| A. pisum | 12 United Kingdom clones | — | 2.4—16.2* |
| N. A. Moran lab clone 5A (Madison, WI) | 0.6 | 4.8 | |
| Diuraphis noxia | P. Baumann lab clone (Lincoln, NE) | 0.9† | 1.8‡ |
| South Africa population | 0.3 | 0.4 | |
| Rhopalosiphum maidis | N. A. Moran lab clone (Tucson) | — | 0.3 |
| S. graminum | Biotype B (K. A. Shufran lab clone) | — | 0.5 |
| Biotype E (T. Mittler lab clone) | 23.5† | 14.5§ | |
| Biotype E (P. Baumann lab clone) | 1.4 | 2.1 | |
| Biotype E (N. A. Moran lab clone) | 1.9 | 1.5 | |
| Biotype E (K. A. Shufran lab clone) | 1.6 | 2.6 | |
| Biotype G (K. A. Shufran lab clone) | 0.5 | 2.4 | |
| Biotype SC (K. A. Shufran lab clone) | — | 0.5 | |
| Uroleucon ambrosiae | 86 individuals, 15 U.S. populations | 0.5—2.8¶ | 0.3—1.9¶ |
Unless otherwise noted, values are previously unpublished and were estimated by using real-time quantitative PCR as in ref. 25 (primer sequences are available upon request). The reason for the discrepancy among Buchnera (S. graminum) estimates is uncertain, although the concordance of the quantitative PCR estimates across biotypes provides support for their accuracy.
Ref. 27; calculated by using quantitative DNA hybridization
Ref. 26; calculated by using quantitative DNA hybridization
Ref. 30; calculated by using quantitative DNA hybridization
Ref. 18; calculated by using quantitative DNA hybridization
Ref. 25; calculated by using real-time quantitative PCR
In the case of trpEG, the variation in ratio of plasmid-borne to chromosomal genes is due both to differences in numbers of repeats per plasmid (21) and to differences in relative plasmid copy number. But another variable affecting the number of functional trpEG copies is the frequent inactivation of some gene copies. Early stop codons, large deletions, and frameshift mutations have been found in at least some of the plasmid-borne trpEG copies of several aphid species (13, 28–31) and can be geographically widespread within a species (28).
The rarity of origins of plasmid amplification, the apparent heritability and stability of amplification levels, and the frequent inactivation of trpEG all suggest that levels of plasmid amplification do not provide a route for quick responses to changes in demands for leucine or tryptophan production. Thus, although the plasmids are likely to be involved in adjusting production of amino acids, the exact mechanism is not clear. This picture is supported by data suggesting that tryptophan production is not correlated with the ratio of trpEG to single copy chromosomal gene copies among different A. pisum strains (27), although these ratios could have included inactivated trpEG copies (see 32). Observations so far do not exclude the possibility of some feedback control of plasmid copy number by amino acid concentrations, as a major heritable component does not rule out plasticity in the ratio of plasmid-borne to chromosomal genes.
Degradation of Symbiont Provisioning Through Gene Inactivation and Loss
In contrast to the situation in A. pisum in which all of the essential amino acids pathways are conserved, several lines of evidence indicate that individual pathways are sometimes inactivated or lost in particular Buchnera strains. The absence of a gene can only be definitively concluded when whole genome sequences are determined. Among the three Buchnera genomes now available for distantly related aphid species with very different patterns of host plant use, loss of the pathway for fixation of inorganic sulfur occurred independently in two lineages; this loss precludes synthesis of cysteine and methonine by using inorganic sulfur sources (11, 12). In addition, Buchnera of Baizongia pistaciae has lost capacity for arginine biosynthesis, with the relevant genes deleted from positions at several locations in the genome (12). The fact that two of the three sequenced Buchnera genomes have lost functionality of some nutrition-related pathways strongly suggests that such losses are widespread evolutionary events. This hypothesis is reinforced by nutritional studies suggesting that certain amino acids are dietary requirements for some aphid/Buchnera isolates. Results of nutritional studies on A. pisum and several other aphid species have been interpreted as indicating species-specific or strain-specific requirements for most of the 10 essential amino acids (including arginine, histidine, isoleucine, leucine, lysine, phenylalanine, threonine, tryptophan, and valine) (e.g., refs. 33 and 34). Nutritional studies also have shown variation among species in whether inorganic sulfate could replace either or both of the sulfur-containing amino acids (e.g., ref. 35).
Genomic sequence data show that the essential amino acid pathways are generally present in Buchnera but sometimes inactivated or lost (11, 12), providing striking support for the following interpretation of nutritional results. First, the usual absence of dietary requirement for essential amino acids, which contrasts with animals generally, reflects provisioning by Buchnera. Second, individual pathways are repeatedly lost in individual Buchnera strains, resulting in reversion to host dependence on dietary sources. Because the Buchnera of most aphids appear to have retained a large complement of amino acid biosynthetic pathways, a corollary is that the lineages undergoing these inactivation events may be evolutionary dead ends. The report of substantially smaller genomes in the Buchnera of some aphid species (36) raises the possibility that even more erosion of biosynthetic capabilities has occurred in some lineages, which may encounter enriched diets or rely on additional, novel symbionts (or “secondary” symbionts) (e.g., refs. 37–40) for some amino acids.
The emerging picture is that provisioning services of obligate symbionts can be eroded by losses of gene function. The lack of recombination or gene uptake by Buchnera implies that such losses are irreversible. Inactivation of genes underlying biosynthetic abilities may occur under circumstances in which dietary supplies are sufficient, and data on phloem sap contents indicates that particular amino acids may be adequate in some plants (4, 41). Nonetheless, these losses are expected to impose permanent limitations on future abilities of aphid lineages to colonize novel foodplants. Possibly, associations with secondary symbionts may be driven by the need to replace functions formerly provided by Buchnera.
Integration of Symbiont Gene Expression with the Nutritional Economy of Hosts
Experiments based on full genome microarrays provide a general overview of patterns of gene expression in a symbiont genome, allowing us to ask questions of how expression diverges from that of related free-living bacteria and to address the issue of whether symbionts are able to alter gene expression to fit host needs. Laboratory and statistical methods underlying the microarray results presented here have been described (42), and we limit ourselves to a generalized overview of symbiont gene expression patterns. Fig. 1B contrasts transcript abundances, grouped by gene functional categories, between Buchnera of Schizaphis graminum and E. coli. Buchnera is represented as samples derived from aphids feeding on nutritionally favorable and unfavorable plants (7); E. coli is represented as cultures from growth media with and without preformed amino acids (J. D. Glasner, personal communication). For both organisms, the profiles across the functional categories are strikingly similar for genome content (Fig. 1 A) versus transcriptome content (Fig. 1B). The fraction of genes in a particular clusters of orthologous groups (COG) category shows a highly significant linear relationship to the pooled expression level for that category, both for Buchnera (for both treatments, R2 = 0.98, P < 0.0001) and E. coli (for both treatments, R2 = 0.93, P < 0.0001). As noted above, the Buchnera genome has differentially retained genes for essential amino acid biosynthesis (a subset of the amino acid metabolism category found in E. coli as shown in Fig. 1), and this greater representation extends to the transcriptome, suggesting greater investment in functions that underlie host provisioning. Another notable feature is that chaperonins are relatively highly expressed in Buchnera, even under nonstress conditions, as previously noted (Fig. 1B and ref. 42).
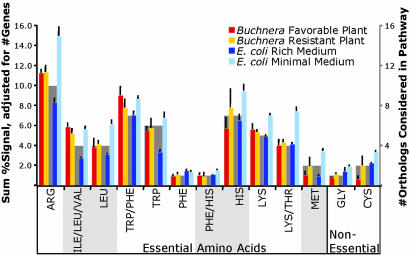
The parallel between number of genes and transcript abundance extends to the level of individual amino acids pathways for both Buchnera and E. coli, although, as expected, the growth media has a major influence on the expression levels in E. coli (Fig. 2). The main amino acids that are deficient in the S. graminum diet (the phloem sap of grasses) are arginine and lysine, which are two of the most common amino acids in the insect proteins (4). Transcript abundances for genes in these pathways tend to be high (Fig. 2). In contrast, genes underlying production of methionine and cysteine show relatively low transcript levels, possibly reflecting the fact that these pathways are of no use for amino acid provisioning in this Buchnera species, because the upstream pathway for sulfur fixation has been inactivated (11).
Fig. 2.
Transcript abundances of genes in essential and nonessential amino acid biosynthetic pathways. Colored bars and left y axis are average transcript signals summed for a given pathway and averaged over experimental replicates. Gray bars and right y axis show numbers of genes per pathway grouping and represent expected signal intensity based on numbers of genes. All data are for orthologs shared between Buchnera(Sg) and E. coli. ARG, argA, argB, argC, argD, argE, carA, carB, argF, argG, and argH; ILE/LEU/VAL, ilvH, ilvI, ilvC, and ilvD; LEU, leuA, leuC, leuD, and leuB; TRP/PHE, aroH, aroB, aroD, aroE, aroK, aroA, and aroC; TRP, trpE, trpD, trpC, trpA, and trpB; PHE, pheA; PHE/HIS, hisC; HIS, hisG, hisI, hisA, hisH, hisF, hisB, and hisD; LYS, dapA, dapB, dapD, dapF, and lysA; LYS/THR, thrA, asd, thrB, and thrC; MET, metE, and metF; GLY, glyA; CYS, cysE, and cysK. Block shading below the graph groups amino acid pathways as branched, aromatic, etc.
As noted above, aphids feed on heterogeneous populations of plants that impose varying demands on Buchnera's biosynthetic capabilities, raising the question of how the symbiont genome responds. In free-living bacteria, availability of a particular amino acid has a dramatic effect on its rate of production, and this feedback control arises from a myriad of mechanisms, involving transcript production as well as inhibition of enzymes. For the relatively long and energetically expensive pathways underlying production of the essential amino acids, transcript production is typically governed by multiple regulatory mechanisms in E. coli and other bacteria. These same pathways underlie the central contribution of Buchnera to host nutrition. In view of the varied diets of hosts and the apparent cost of excess production of biosynthetic enzymes and end products, we might expect mechanisms for transcriptional regulation of these genes in Buchnera. This expectation is reinforced by the fact that amino acid provisioning comprises a relatively large proportion of the genome and transcriptome of Buchnera.
But, if transcriptional control mechanisms are present at all, they are altered dramatically in Buchnera. The first evidence for loss of the usual regulatory mechanisms came from examination of regions upstream to structural genes for the biosynthetic enzymes. These lack leader peptides and show altered sequences that indicate degraded or altered binding properties, as noted, for example, for aromatic amino acid pathways (43, 44). The first complete genome of Buchnera revealed the lack of recognizable orthologs of any of the regulatory proteins that bind with free amino acids to block or activate transcriptional promoter sites (13), although metR was identified as an intact ORF in the second genome (11).
The loss of recognizable gene regulatory features in sequence analyses constrasts with findings of physiological studies. Several of these have provided evidence that aphids with an intact symbiosis can down-regulate their production of specific amino acids when abundant. This suggests that Buchnera has one or more negative feedback mechanisms governing amino acid production (45–47). For example, in studies on Aphis fabae provided with radioactively labeled glutamic acid, a greater proportion of label was incorporated into the essential amino acid isoleucine on diets lacking isoleucine than on diets containing it (48). Similarly, in A. pisum, neosynthesis of all essential amino acids was depressed on a diet with plentiful essential amino acids relative to diets with limiting amounts, with synthesis of histidine and arginine completely suppressed (45). Although it is plausible that the apparent mechanisms for regulating amino acid production involve only feedback inhibition of the enzymes rather than transcriptional regulation, this would provide a stark contrast to the situation in other bacteria, in which regulation of transcription is a prominent means of control.
As reported in Wei et al. (49), the fraction of the E. coli transcriptome devoted to amino acid biosynthetic genes increases markedly for colonies grown on minimal media (no exogenous amino acids provided). For Buchnera, in contrast, pooled transcript abundance for genes underlying amino acid biosynthesis differs little from aphids confined to plants with dramatically different nutritional qualities (Fig. 1B). This relative constancy of transcript abundances extends to individual biosynthetic pathways (Fig. 2). The low response to changes in plant quality could reflect a lack of symbiont ability to adaptively alter transcription rates. However, other explanations are possible, including experimental limitations preventing detection of fine-tuned feedback mechanisms; for example, the pooling of RNA from maternal and embryonic symbionts may obscure biologically meaningful changes. Regulation of amino acid biosynthesis could be achieved, in part, through regulation of amino acid transport from host to symbiont. The Buchnera genome lacks apparent importers, but each Buchnera cell is enclosed within a host membrane that is capable of active transport of glutamate and aspartate, which are substrates for biosynthesis of the other amino acids (10). Such host control could govern the overall levels of amino acid biosynthesis within Buchnera cells but would not provide a way to fine-tune the production of individual amino acids according to host needs. The aphids seem not to alter their assimilation of essential amino acids in response to different dietary concentrations of these compounds (50), indicating that their regulation within aphids must involve differential symbiont productivity.
Conclusions
The genomes of the obligate symbionts of insects provide a clear picture of modification of an ancestral genome for a particular role in exchanging nutrients with the host. The picture emerging so far tells us that the essential genetic capabilities are ancient and consist of pathways that are widely distributed among nonsymbiotic bacteria. It appears that the maternally inherited, obligate bacteriome-associates of insects are derived through reduction and minor modifications of ancestral genomes rather than through acquisition or invention of novel “symbiosis” genes. Nonetheless, some of the critical elements for maintenance of symbiosis remain unidentified. We do not yet know why the γ3 Proteobacteria have given rise to a large proportion of the symbiotic lineages living in insects. Perhaps this group possesses some yet-to-be-discovered key genes that enable symbiosis. One possibility is the frequent presence of a type III secretion apparatus that might serve as the initiator of the intracellular association, as in the secondary symbionts of tsetse flies and the related symbionts of grain weevils (51). In addition, the control of gene expression may involve novel mechanisms, a possibility that is supported by the apparent plasticity of Buchnera contributions depending on host diet.
Acknowledgments
We thank H. McLaughlin and H. Dunbar for laboratory assistance, K. A. Shufran for providing aphid clones, J. Glasner for access to unpublished data, and H. Ochman for comments on the manuscript. Funding was from National Science Foundation Grant 9978518 (to N.A.M.), a National Science Foundation postdoctoral fellowship (to J.L.W.), and a National Institutes of Health postdoctoral fellowship (to G.R.P.).
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Chemical Communication in a Post-Genomic World,” held January 17–19, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
References
- 1.Moran, N. A. (2002) Cell 108, 583-586. [DOI] [PubMed] [Google Scholar]
- 2.Buchner, P. (1965) Endosymbiosis of Animals with Plant Microorganisms (Wiley, New York).
- 3.Munson, M. A., Baumann, P., Clark, M. A., Baumann, L., Moran, N. A., Voegtlin, D. J. & Campbell, B. C. (1991) J. Bacteriol. 173, 6321-6324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sandström, J. P. & Moran, N. A. (2001) Physiol. Entomol. 26, 202-211. [Google Scholar]
- 5.Bernays, E. A. & Klein, B. A. (2002) Physiol. Entomol. 27, 275-284. [Google Scholar]
- 6.Sandström, J. & Pettersson, J. (1994) J. Insect Physiol. 40, 947-955. [Google Scholar]
- 7.Telang, A., Sandström, J., Dyreson, E. & Moran, N. A. (1999) Entomol. Exp. Appl. 91, 402-412. [Google Scholar]
- 8.Febvay, G., Liadouze, I., Guillaud, J. & Bonnot, G. (1995) Arch. Insect Biochem. Physiol. 29, 45-69. [Google Scholar]
- 9.Sasaki, T. & Ishikawa, H. (1995) J. Insect Physiol. 41, 41-46. [Google Scholar]
- 10.Whitehead, L. F. & Douglas, A. E. (1993) J. Gen. Microbiol. 139, 821-826. [Google Scholar]
- 11.Tamas, I., Klasson, L., Canback, B., Naslund, A. K., Eriksson, A. S., Wernegreen, J. J., Sandström, J. P., Moran, N. A. & Andersson, S. G. (2002) Science 296, 2376-2379. [DOI] [PubMed] [Google Scholar]
- 12.van Ham, R. C., Kamerbeek, J., Palacios, C., Rausell, C., Abascal, F., Bastolla, U., Fernández, J. M., Jiménez, L., Postigo, M., Silva, F. J., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 581-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shigenobu, S., Watanabe, H., Hattori, M., Sakaki, Y. & Ishikawa, H. (2000) Nature 407, 81-86. [DOI] [PubMed] [Google Scholar]
- 14.Akman, L., Yamashita, A., Watanabe, H., Oshima, K., Shiba, T., Hattori, M. & Aksoy, S. (2002) Nat. Genet. 32, 402-407. [DOI] [PubMed] [Google Scholar]
- 15.Gil, R., Silva, F. J., Zientz, E., Delmotte, F., Gonzalez-Candelas, F., Latorre, A., Rausell, C., Kamerbeek, J., Gadau, J., Holldobler, B., et al. (2003) Proc. Natl. Acad. Sci. USA 100, 9388-9393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moran, N. A. & Mira, A. (2001) Genome Biol. 2, RESEARCH0054. [DOI] [PMC free article] [PubMed]
- 17.Silva, F. J., Latorre, A. & Moya, A. (2001) Trends Genet. 17, 615-618. [DOI] [PubMed] [Google Scholar]
- 18.Lai, C. Y., Baumann, L. & Baumann, P. (1994) Proc. Natl. Acad. Sci. USA 91, 3819-3823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bracho, A. M., Martinez-Torres, D., Moya, A. & Latorre, A. (1995) J. Mol. Evol. 41, 67-73. [DOI] [PubMed] [Google Scholar]
- 20.Baumann, L., Baumann, P., Moran, N. A., Sandström, J. & Thao, M. L. (1999) J. Mol. Evol. 48, 77-85. [DOI] [PubMed] [Google Scholar]
- 21.Rouhbakhsh, D., Clark, M. A., Baumann, L., Moran, N. A. & Baumann, P. (1997) Mol. Phylogenet. Evol. 8, 167-176. [DOI] [PubMed] [Google Scholar]
- 22.van Ham, R. C., González-Candelas, F., Silva, F. J., Sabater, B., Moya, A. & Latorre, A. (2000) Proc. Natl. Acad. Sci. USA 97, 10855-10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lai, C. Y., Baumann, P. & Moran, N. A. (1995) Insect Mol. Biol. 4, 47-59. [DOI] [PubMed] [Google Scholar]
- 24.Komaki, K. & Ishikawa, H. (1999) J. Mol. Evol. 48, 717-722. [DOI] [PubMed] [Google Scholar]
- 25.Plague, G. R., Dale, C. & Moran, N. A. (2003) Mol. Ecol. 12, 1095-1100. [DOI] [PubMed] [Google Scholar]
- 26.Thao, M. L., Baumann, L., Baumann, P. & Moran, N. A. (1998) Curr. Microbiol. 36, 238-240. [DOI] [PubMed] [Google Scholar]
- 27.Birkle, L. M., Minto, L. B. & Douglas, A. E. (2002) Physiol. Entomol, 27, 302-306. [Google Scholar]
- 28.Wernegreen, J. J. & Moran, N. A. (2000) Proc. R. Soc. London Ser. B 267, 1423-1431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Baumann, L., Clark, M. A., Rouhbakhsh, D., Baumann, P., Moran, N. A. & Voegtlin, D. J. (1997) Curr. Microbiol. 35, 18-21. [Google Scholar]
- 30.Lai, C. Y., Baumann, P. & Moran, N. A. (1996) Appl. Environ. Microbiol. 62, 332-339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wernegreen, J. J., Richardson, A. O. & Moran, N. A. (2001) Mol. Phylogenet. Evol. 19, 479-485. [DOI] [PubMed] [Google Scholar]
- 32.Birkle, L. M. & Douglas, A. E. (1999) Heredity 82, 605-612. [DOI] [PubMed] [Google Scholar]
- 33.Srivastava, P. N. & Auclair, J. L. (1975) J. Insect Physiol. 21, 1865-1871. [Google Scholar]
- 34.Srivastava, P. N. (1987) in World Crop Pests, eds. Mink, A. K. & Harrewijn, P. (Elsevier, Amsterdam), Vol. 2A, pp. 99-122. [Google Scholar]
- 35.Turner, R. B. (1971) J. Insect Physiol. 17, 2451-2456. [Google Scholar]
- 36.Gil, R., Sabater-Muñoz, B., Latorre, A., Silva, F. J. & Moya, A. (2002) Proc. Natl. Acad. Sci. USA 99, 4454-4458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Fukatsu, T. (2001) Appl. Environ. Microbiol. 67, 5315-5320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Montllor, C. B., Maxmen, A. & Purcell, A. H. (2002) Ecol. Entomol. 27, 189-195. [Google Scholar]
- 39.Sandström, J. P., Russell, J. A., White, J. P. & Moran, N. A. (2001) Mol. Ecol. 10, 217-228. [DOI] [PubMed] [Google Scholar]
- 40.Tsuchida, T., Koga, R., Shibao, H., Matsumoto, T. & Fukatsu, T. (2002) Mol. Ecol. 11, 2123-2135. [DOI] [PubMed] [Google Scholar]
- 41.Sandström, J. & Moran, N. A. (1999) Ent. Exp. Appl. 91, 203-210. [Google Scholar]
- 42.Wilcox, J. L., Dunbar, H. E., Wolfinger, R. D. & Moran, N. A. (2003) Mol. Microbiol. 48, 1491-1500. [DOI] [PubMed] [Google Scholar]
- 43.Jimenez, N., González-Candelas, F. & Silva, F. J. (2000) J. Bacteriol. 182, 2967-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kolibachuk, D., Rouhbakhsh, D. & Baumann, P. (1995) Curr. Microbiol. 30, 313-316. [DOI] [PubMed] [Google Scholar]
- 45.Febvay, G., Rahbé, Y., Rynkiewiecz, M., Guillard, J. & Bonnot, G. (1999) J. Exp. Biol. 202, 2639-2652. [DOI] [PubMed] [Google Scholar]
- 46.Leckstein, P. M. & Llewellyn, M. (1973) J. Insect Physiol. 19, 973-980. [DOI] [PubMed] [Google Scholar]
- 47.Liadouze, I., Febvay, G., Guillaud, J. & Bonnot, G. (1995) J. Insect Physiol. 41, 33-40. [Google Scholar]
- 48.Douglas, A. E., Minto, L. B. & Wilkinson, T. L. (2001) J. Exp. Biol. 204, 349-358. [DOI] [PubMed] [Google Scholar]
- 49.Wei, Y., Lee, Y. M., Richmond, C., Blattner, F. R., Rafalski, J. A. & LaRossa, R. A. (2001) J. Bacteriol. 183, 545-556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wilkinson, T. L. & Ishikawa, H. (1999) Entomol. Exp. Appl. 91, 195-201. [Google Scholar]
- 51.Dale, C., Plague, G. R., Wang, B., Ochman, H. & Moran, N. A. (2002) Proc. Natl. Acad. Sci. USA 99, 12397-12402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Tatusov, R. L., Natale, D. A., Garkavtsev, I. V., Tatusova, T. A., Shankavaram, U. T., Rao, B. S., Kiryutin, B., Galperin, M. Y., Fedorova, N. D. & Koonin, E. V. (2001) Nucleic Acids Res. 29, 22-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Glasner, J. D., Liss, P., Plunkett, G., III, Darling, A., Prasad, T., Rusch, M., Byrnes, A., Gilson, M., Biehl, B., Blattner, F. R. & Perna, N. T. (2003) Nucleic Acids Res. 31, 147-151. [DOI] [PMC free article] [PubMed] [Google Scholar]