Abstract
Cell–cell communication in bacteria is accomplished through the exchange of chemical signal molecules called autoinducers. This process, called quorum sensing, allows bacteria to monitor their environment for the presence of other bacteria and to respond to fluctuations in the number and/or species present by altering particular behaviors. Most quorum-sensing systems are species- or group-specific, which presumably prevents confusion in mixed-species environments. However, some quorum-sensing circuits control behaviors that involve interactions among bacterial species. These quorum-sensing circuits can involve both intra- and interspecies communication mechanisms. Finally, anti-quorumsensing strategies are present in both bacteria and eukaryotes, and these are apparently designed to combat bacteria that rely on cell–cell communication for the successful adaptation to particular niches.
Keywords: quorum sensing, autoinducer, cell–cell communication
In a process called quorum sensing, bacteria monitor the presence of other bacteria in their surroundings by producing and responding to signaling molecules known as autoinducers. The concentration of autoinducer in a given environment is proportional to the number of bacteria present; therefore, detecting autoinducers gives bacteria a mechanism for “counting” one another. Responding to autoinducers by altering gene expression gives bacteria a means to perform particular behaviors only when living in a community but not when living in isolation. Most quorum-sensing controlled behaviors are productive only when a group of bacteria carries them out in synchrony; they include bioluminescence, secretion of virulence factors, biofilm formation, sporulation, conjugation, and pigment production (1–3). Because quorum sensing allows bacteria to coordinate the behavior of the group, it enables them to take on some of the characteristics of multicellular organisms.
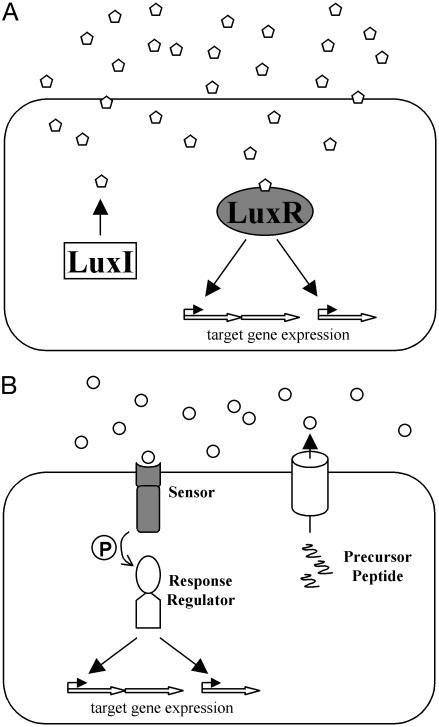
There are two general types of bacterial quorum-sensing systems: Gram-negative LuxIR circuits and Gram-positive oligopeptide two-component circuits. Gram-negative quorumsensing bacteria typically possess proteins homologous to the LuxI and LuxR proteins of Vibrio fischeri, the bacterium in which they were initially discovered (Fig. 1A) (4). The LuxI-type proteins catalyze the formation of a specific acyl-homoserine lactone (AHL) autoinducer that freely diffuses into and out of the cell and increases in concentration in proportion to cell population density. The LuxR-type proteins each bind a specific AHL autoinducer when the concentration of autoinducer reaches a threshold level. The LuxR–AHL complexes activate transcription of target genes by recognizing and binding specific DNA sequences at quorum-sensing-regulated promoters (4–6). Some functions controlled by LuxIR-type quorum-sensing systems include plasmid conjugation in Agrobacterium tumefaciens (7), antibiotic production in Erwinia carotovora (8), biofilm production and virulence gene expression in Pseudomonas aeruginosa (9, 10), and expression of factors necessary for symbiosis in Sinorhizobium meliloti (11). Currently, there are >70 known LuxIR quorum-sensing systems in Gram-negative bacteria (1, 12–14).
Fig. 1.
Canonical quorum-sensing circuits. (A) In typical Gram-negative LuxIR circuits, the LuxI-type protein catalyzes the synthesis of an AHL autoinducer (pentagons). The LuxR-type protein binds the AHL and controls the expression of target genes. (B) In typical Gram-positive AIP two-component quorumsensing circuits, precursor peptides are cleaved, modified, and exported by dedicated transporters. The resulting AIPs (circles) are detected by twocomponent sensor-histidine kinases, and sensory information is relayed by phosphorylation (P) of cognate response regulators that, in turn, control target gene expression. The proteins responsible for autoinducer binding are shaded in gray.
Low G+C Gram-positive bacteria typically use modified oligopeptides as autoinducers (15–17). These signals are generically referred to as autoinducing polypeptides (AIPs) (Fig. 1B). AIPs are produced in the cytoplasm as precursor peptides and are subsequently cleaved, modified, and exported. AIPs specifically interact with the external domains of membrane-bound two-component sensor kinase proteins. Interaction of the autoinducer with its cognate sensor stimulates the kinase activity of the sensor kinase protein, resulting in the phosphorylation of its partner response regulator protein. The phosphorylated response regulator protein binds DNA and alters the transcription of target genes. Some examples of behaviors controlled by AIP quorum-sensing systems include genetic competence and sporulation in Bacillus subtilis (18, 19), competence for DNA uptake in Streptococcus pneumoniae (20), and virulence factor expression in Staphylococcus aureus (21) and Enterococcus faecalis (22).
Despite the fact that the autoinducer signals and detection apparatuses can be highly similar, both LuxIR and oligopeptide two-component-type quorum-sensing systems function such that a response is elicited only to the autoinducer of the bacterial species that produced it. This signaling specificity stems in large part from subtle differences in the autoinducer molecules and their receptors. Here we discuss what is known about the requirements for species-specific cell–cell communication. Quorum-sensing circuits also exist that are species nonspecific; their properties are discussed below.
Specificity in AHL Communication Systems
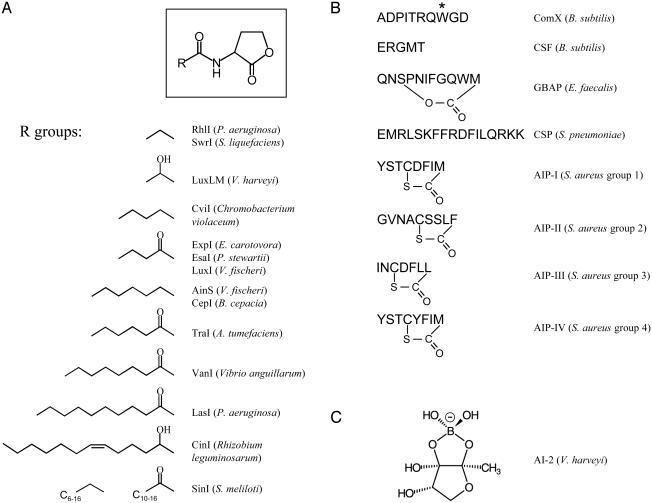
AHL autoinducers all share a common homoserine lactone moiety and differ only in their acyl side chain moieties (Fig. 2A) (3, 12). However, an autoinducer of one bacterial species rarely influences expression of target genes in another species. Two primary factors control this exquisite signaling specificity: first, the substrate specificity of the LuxI-like proteins, and second, specificity in the binding of the LuxR-type proteins to their cognate AHLs.
Fig. 2.
Structures of different autoinducers. (A Upper) AHL autoinducers share a common homoserine lactone moiety. (Lower) Side chains of some different AHLs are shown (R groups). The synthases and the organisms that produce them are listed. (B) AIP peptide sequences, their designations, and the organisms that produce them are shown. The asterisk above the tryptophan residue in ComX represents an isoprenyl group. (C) AI-2 of V. harveyi.
LuxI proteins link the side chain group of specific acyl-acyl carrier proteins to the homocysteine moiety of S-adenosylmethionine (23–25). These reactions are very precise, because LuxI proteins recognize only the ACP containing a specific acyl chain moiety. The reliability of this reaction is vital to maintaining species-specific communication within each quorum-sensing system, because it ensures that each LuxI protein generates only the correct AHL signal molecule.
The molecular interactions underlying the specificity in LuxI-directed signal production have been explored by structural analyses of EsaI, the LuxI homologue of Pantoea stewartii (26). EsaI catalyzes the formation of the AHL 3-oxohexanoylhomoserine lactone. The x-ray crystal structure of EsaI shows that a hydrophobic cavity in the protein likely encapsulates the acyl moiety of the acyl-acyl carrier proteins. The extreme preference of EsaI for a six-carbon acyl side chain is due to the size of the binding pocket. In addition, preference for a 3-oxo AHL results from a favorable hydrogen bond between the C3 carbonyl of the AHL and the hydroxyl group of a threonine residue on EsaI. Modeling studies suggest that the corresponding pocket in LasI of P. aeruginosa is larger, and consistent with this, LasI produces a longer autoinducer, 3-oxododecanoylhomoserine lactone. This study suggests that the length and derivatization of the acyl side chains of AHLs are determined by differences in the structures of the binding cavities of the LuxI-type proteins.
Specificity in LuxR–AHL interaction is critical for bacteria to distinguish AHLs produced by their own species from AHLs of other species. The structural basis for ligand–receptor interaction has been most thoroughly examined with the TraR protein of A. tumefaciens and its cognate AHL autoinducer, 3-oxooctanoyl-homoserine lactone (27). The x-ray structure of TraR in complex with its cognate AHL and DNA shows a precise interaction between TraR and the acyl moiety on the AHL. In this case, the C3 keto group of the AHL is stabilized by hydrogen bonding to a water molecule present in an autoinducer-binding cavity. Presumably, alterations in the size and shape of the AHL-binding pocket in other LuxR proteins will correspond to side chain lengths and substitutions of their cognate ligands.
The specificity of the AHL-LuxR pair does not extend to DNA binding and transcriptional activation, because the LuxR proteins all bind to similar DNA regulatory elements termed “lux boxes” (28, 29). Although interchangeability between AHLs and their LuxR partners is not tolerated, together the pairs can control gene expression at noncognate lux boxes. For example, the AHL molecule produced by V. fischeri (3-oxohexanoylhomoserine lactone) together with LuxR, expressed in Escherichia coli, can induce transcription of lasB, a gene normally regulated by LasR and 3-oxododecanoyl-homoserine lactone in P. aeruginosa. Similarly, 3-oxododecanoyl-homoserine lactone, coupled with LasR and expressed in E. coli, will activate the V. fischeri quorum-sensing controlled target luxCDABE (29). Again, these findings support the hypothesis that target specificity inherent in LuxIR circuits stems exclusively from the selection of a particular AHL by its cognate LuxR-type protein.
Specificity in AIP Communication Systems
As in LuxIR circuits, similarities exist between oligopeptide autoinducers and cognate two-component sensors from different groups or species of Gram-positive bacteria, yet each signal-response system is remarkably specific. AIP synthesis is inherently accurate, because precise signal generation is guaranteed by the specific DNA sequence encoding the precursor protein. All AIPs are cleaved from longer precursor peptides, and many AIPs are also subject to posttranslational modifications (Fig. 2B). Signaling accuracy is achieved through the highly sensitive nature of the AIP receptors to alterations in AIP structure (16).
AIP signaling specificity has been analyzed extensively in S. aureus (30–32). Several clinically important groups within the S. aureus species have been characterized, and they all have slightly different AIPs, processing enzymes, and receptors (33, 34). The AIPs are 8 to 9 aa long and contain thiolactone rings involving invariant cysteine residues situated 5 aa from the C terminus (Fig. 2B) (35). Specificity in AIP signaling, at least in the staphylococci, is entirely determined by the AIP–sensor kinase interaction. This hypothesis is supported by findings that the cognate response regulators in all of the S. aureus groups are completely conserved, as are the target promoter regions (30, 33). Interestingly, the AIP produced by one group of S. aureus not only activates its own virulence cascade but also inhibits virulence in all other groups. Inhibition of virulence requires far less specificity in AIP structure, because the thiolactone ring portion common to all of the S. aureus AIPs acts as a universal inhibitor of AIP signaling (30–32). The group-specific AIP signaling in S. aureus indicates that quorum sensing in S. aureus occurs at the subspecies level. Inhibition by one S. aureus group of cell–cell communication in other S. aureus groups is presumed to benefit the group that first establishes its quorum-sensing cascade, because it facilitates the formation of a single-group infection.
Quorum Sensing Using Multiple Autoinducers and Sensors
Quorum-sensing systems consisting of single autoinducer-sensor pairs are sufficient to control gene expression in response to changes in cell density. However, many bacteria have two or more quorum-sensing systems. Use of multiple quorum-sensing systems allows bacteria to integrate pieces of sensory information, which presumably confers plasticity to the genetic network. The hierarchies can be set up in series or in parallel, the former enabling regulation of genes in a temporally defined manner, and the latter enabling regulation of discrete groups of genes or converging to regulate an identical set of genes.
In P. aeruginosa, two AHL quorum-sensing systems, LasIR and RhlIR, act in series. At high cell density, the concentration of both AHLs is high, and LasR binds its specific AHL to activate the expression of particular target genes. One of the genes activated by the AHL–LasR complex is rhlR, which encodes a second AHL receptor, RhlR. RhlR, in turn, binds its cognate AHL autoinducer and induces expression of its own target genes. Thus, genes controlled by the LasIR system are expressed before those controlled by the RhlIR system. This temporal pattern of gene regulation allows P. aeruginosa to express different virulence factors at various stages in the infection process (36, 37).
The two P. aeruginosa AHL signals promote virulence in a variety of different hosts, the most important human example being the cystic fibrosis (CF) lung. Not only is the CF lung susceptible to damage caused by AHL-controlled virulence factors, but it is also vulnerable to direct effects of the AHL molecules themselves. Specifically, the P. aeruginosa autoinducers induce the production of the chemokine IL-8 in the CF lung. As a result, neutrophils are recruited to the lung, and this action facilitates the formation of a potent P. aeruginosa infection. Thus, in this system, bacterial AHLs direct the expression of bacterial and host factors that enhance the infection process (38).
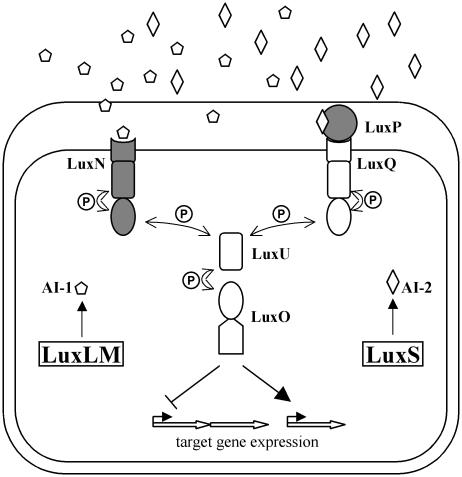
In two Vibrio species, Vibrio harveyi and Vibrio cholerae, multiple quorum-sensing systems converge to regulate a single group of genes. V. harveyi uses two parallel systems to regulate the expression of target genes, including those required for bioluminescence, and V. cholerae has three systems that jointly control the virulence regulon (1, 39). In V. harveyi (Fig. 3), the two autoinducer synthases, LuxLM and LuxS, each catalyze the synthesis of a specific autoinducer: the AHL 3-hydroxybutanoylhomoserine lactone (denoted AI-1) in the case of LuxLM, and a unique furanosyl borate diester, 3a-methyl-5,6-dihydrofuro-[2,3-d][1,3,2]dioxaborole-2,2,6,6a-tetraol (AI-2), in the case of LuxS (Fig. 2 A and C) (40–42). Unlike other Gram-negative quorum-sensing systems in which AHL autoinducers are detected by a cytoplasmic LuxR-type protein, detection of the two autoinducers in V. harveyi occurs in the periplasm via cognate two-component sensor kinase proteins. AI-1 initiates signal transduction in the sensor kinase protein LuxN (43), and AI-2 binds the periplasmic binding protein LuxP, which in turn initiates signaling from the sensor kinase protein LuxQ (44). Information from both LuxN and LuxPQ converges at the phosphorelay protein LuxU (45, 46), and LuxU transmits the phosphorylation signal to the response regulator LuxO. LuxO controls transcription of target genes including those encoding luciferase (luxCDABE) (47, 48).
Fig. 3.
The V. harveyi quorum-sensing circuit. Two autoinducers, the AHL AI-1 (pentagons) and AI-2 (diamonds), are produced by their cognate synthases LuxLM and LuxS, respectively. AI-1 and AI-2 bind cognate sensors (LuxN and LuxPQ, respectively), initiating a phosphorylation cascade (P) that travels through LuxU and alters the phosphorylation state of the response regulator protein LuxO. LuxO controls the expression of target genes. The proteins responsible for autoinducer binding are shaded in gray.
It seems paradoxical that V. harveyi uses two quorum-sensing systems to regulate the same set of target genes, because either system alone should be sufficient. One possible explanation for this molecular setup is that the autoinducers play different roles in cell–cell communication. Support for this idea stems from the finding that AI-1, like other AHL autoinducers, is species-specific, whereas many diverse species of bacteria possess a conserved LuxS homologue and produce AI-2, suggesting that AI-2 may function in interspecies cell–cell communication. Additionally, unlike AHL and AIP signaling, which is restricted to Gram-negatives and -positives, respectively, LuxS and AI-2 exist in both Gram-negative and -positive bacteria, suggesting that AI-2-mediated communication arose before AHL and AIP signaling (2, 42, 49).
The V. harveyi quorum-sensing circuit can distinguish among the presence of none, one, or both autoinducers. However, maximal expression of Lux occurs only when both autoinducers are present, suggesting that the circuit could act as a coincidence detector for both autoinducers (50). Because the V. harveyi quorum-sensing regulon has not yet been fully defined, it remains possible that additional classes of target genes exist that are regulated exclusively by either AI-1 or AI-2. If so, V. harveyi could differentially modify its behavior depending on whether it makes up the majority or the minority of a given mixed-species population. If, on the other hand, the V. harveyi quorum-sensing circuit functions primarily as a coincidence detector that regulates gene expression in response to the presence of both autoinducers, the advantage of this scheme could be that the simultaneous detection of two signals reduces the vulnerability of the circuit to noise or to “trickery.” A system that works by a combinatorial scheme could protect the cell–cell communication circuit from molecules made by other organisms in the environment that are similar in structure to the autoinducers (50).
Interspecies Cell–Cell Communication: LuxS and AI-2
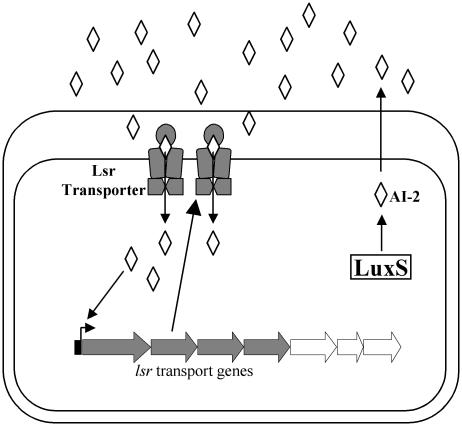
AI-2 is the only species-nonspecific autoinducer known. Because of its widespread occurrence, AI-2 is proposed to act as a universal quorum-sensing signal for interaction between species of bacteria (42, 49). Besides controlling light production in V. harveyi, the various roles that AI-2 plays in other species are beginning to be defined. Among other things, AI-2 controls the expression of genes required for virulence in E. coli, V. cholerae, Clostridium perfringens, and Streptococcus pyogenes; iron acquisition in Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans; antibiotic production in Photorhabdus luminescens; motility in Campylobacter jejuni; and mixed-species biofilm formation between P. gingivalis and Streptococcus gordonii (51). In Salmonella typhimurium, AI-2 was recently shown to control a seven-gene operon, called the lsr operon (for LuxS Regulated) (Fig. 4) (52). Four of the lsr operon genes encode an ABC transporter whose function is to promote internalization of AI-2. No additional AI-2-regulated genes have been identified in S. typhimurium, suggesting that AI-2 may have a role that is different from a classic quorum-sensing autoinducer in some bacteria, including S. typhimurium.
Fig. 4.
Lsr-mediated transport of AI-2 in S. typhimurium. AI-2 activates transcription of the lsr genes, four of which (shaded in gray) encode the Lsr transporter apparatus that functions to internalize AI-2.
In S. typhimurium, and presumably in other bacteria that possess the lsr operon, AI-2 is produced in the cytoplasm and is released into the extracellular environment, where it accumulates. AI-2 is subsequently transported back inside the cell through the Lsr transporter (Fig. 4) (52). The benefit that bacteria gain from this cyclic process is unknown, although many possibilities exist. For example, AI-2 could be a quorum-sensing signal that impacts as-yet-unidentified genes in S. typhimurium. Other quorum-sensing regulated genes may exist that were not identified in the analysis revealing the lsr genes due to redundancy in the quorum-sensing circuitry or a requirement for a specific growth condition that was not met in the laboratory. If so, import of AI-2 could be used to terminate the quorumsensing cascade or to convey AI-2 to an internal detection apparatus. An alternative hypothesis is that internalization of AI-2 could be a mechanism that bacteria such as S. typhimurium have evolved to interfere with the AI-2 quorum-sensing systems of competing species. This action could serve to confound other bacteria that regulate specific functions using AI-2, thereby giving S. typhimurium an advantage in particular environments.
Interference with Cell–Cell Communication: Biological Battles and Conspiracies
Quorum sensing often promotes behaviors that are detrimental to other organisms in the vicinity. For example, pathogens such as P. aeruginosa and E. carotovora use AHL-mediated quorum sensing to activate their virulence genes in specific animal and plant hosts, respectively (12). Likewise, in Serratia liquefaciens, antibiotic production and swarming motility, two behaviors required for virulence, are controlled by AHL-mediated quorum sensing (53). To protect themselves, some susceptible organisms have developed defenses that interfere with quorum sensing. Several anti-AHL-mediated quorum-sensing strategies have recently been discovered, and we present a few examples here. Additionally, in mixed-species environments, different species of bacteria can “conspire” with one another by responding to heterologous AHLs.
The macroalga Delisea pulchra produces a collection of halogenated furanones, many of which inhibit growth of pathogenic microorganisms on its surface. The furanones have structural similarity to AHLs and can prevent the AHL signal of S. liquefaciens from interacting with its cognate LuxR-type protein. In this model system, inhibition of AHL signaling prevents AHL-mediated motility in S. liquefaciens, which precludes bacterial colonization of the alga (54–57). Similarly, several organisms produce enzymes that modify AHLs, rendering them inactive. Marine algae such as Laminaria digitata prevent the formation of bacterial biofilms on their surfaces by producing haloperoxidases that generate oxidized halogens with microbicidal activity, such as hypochlorous acid (HOCl). Oxidized halogens react specifically with C3-oxo-AHLs and destroy their signaling capability. This finding is particularly significant because of the ability of the oxidized halogens to penetrate into biofilms (58).
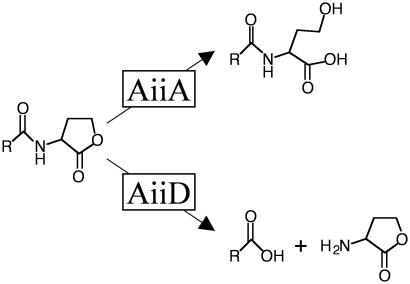
In addition to eukaryotes, some bacteria produce enzymes that interfere with AHL signaling, presumably to gain an advantage over AHL-producing bacteria in specific biological niches. For example, an AHL lactonase (AiiA) isolated from Bacillus species has been shown to hydrolyze the lactone ring of AHLs to form acylhomoserine, which does not function as a quorum-sensing signal (Fig. 5 Upper). Expression of AiiA in either the quorum-sensing pathogen E. carotovora or its susceptible plant host eliminates AHL production and infection, suggesting that AiiA can be used to prevent infection by pathogens that use quorum sensing to control virulence (59–62). What in vivo role AiiA plays in Bacillus species is not known, but it could be used to interfere with AHL-mediated quorum-sensing behaviors of soil bacteria that compete with Bacillus for the same niche. A different AHL inactivating enzyme, AiiD, was recently discovered in a Ralstonia strain isolated from a bacterial biofilm. Like AiiA, AiiD hydrolyzes AHLs, but in this case the side chain is released from the intact homoserine lactone ring (Fig. 5 Lower). Again, the final result is elimination of quorum sensing. Introduction of aiiD into P. aeruginosa caused reduced accumulation of extracellular AHLs and inhibited AHL regulated behaviors such as swarming motility, virulence factor production, and paralysis of Caenorhabditis elegans (63). In another study, the bacterium Variovorax paradoxus was shown to be capable of using AHLs as a sole carbon and nitrogen source, suggesting that it too has AHL degrading activity (64). It appears that V. paradoxus has evolved the ability to degrade AHLs either as a defense against other bacteria that use AHLs as signaling molecules or as a metabolic scavenging method for using molecules released by neighboring bacteria. These recent discoveries suggest that the inhibition of AHL-mediated quorum sensing might be a widespread mechanism that bacteria use to compete with one another.
Fig. 5.
Enzymatic inactivation of AHLs. (Upper) AiiA of Bacillus species hydrolyzes the lactone ring of AHLs. (Lower) AiiD of a Ralstonia strain hydrolyzes AHLs to release the acyl side chain moiety and homoserine lactone.
Instead of interspecies inhibition of AHL signaling, some bacteria use AHLs for interspecies communication during infection. For example, in cystic fibrosis lungs colonized by both P. aeruginosa and Burkholderia cepacia, in addition to producing and responding to its own AHL, B. cepacia responds to AHLs produced by P. aeruginosa, which promotes formation of a mixed-species biofilm (65). Because most bacteria reside in mixed-species environments, other examples of bacteria “conspiring” to carry out different behaviors could be discovered in examinations of quorum sensing in mixed populations.
Few cases of interference with AIP-mediated quorum-sensing systems are currently known, with the best-studied example being the cross-group inhibition of AIP signaling in S. aureus, discussed above. However, other systems for interfering with AIP-mediated quorum-sensing systems may exist. Although the autoinducer AI-2 is used for interspecies communication, no cases of interference with AI-2 quorum sensing are yet documented. However, as noted above, internalization of AI-2 by enterics such as S. typhimurium is considered one such possible anti-AI-2 communication mechanism.
Conclusion
Recent research shows that chemical communication among bacteria is widespread and involves complex interconnected regulatory networks that serve to fine-tune the expression of diverse group behaviors. Specificity in autoinducer production and recognition is a key component of quorum sensing, because bacteria must preserve the fidelity of their communication circuits while existing in communities containing other organisms that produce molecules similar to their own autoinducers, either as quorum-sensing autoinducers or as autoinducer antagonists. The use of quorum sensing to control behaviors such as biofilm formation, symbiosis, and virulence factor expression indicates that quorum sensing is frequently used to regulate traits involving interactions between different organisms, both mutualistic and antagonistic. Thus, the ability of bacteria to distinguish self from other could also be a fundamental property of quorum-sensing systems. In support of this idea, it appears that some bacteria have evolved species-specific as well as “generic” signaling molecules. The recent discovery of autoinducer degrading enzymes demonstrates that interference with quorum sensing is an effective antibacterial strategy used in the wild. Synthetic anti-quorum-sensing strategies could be developed in the future as possible alternatives to antibiotics, because blocking cell–cell communication within or among bacterial species could prevent pathogenicity. Likewise, biotechnological approaches that promote beneficial quorum-sensing behaviors could be exploited for the production of industrial-scale natural products in bacteria.
Acknowledgments
This work was supported by Office of Naval Research Grant N00014-99-1-0767, National Science Foundation Grant MCB-0094447, and National Institute of General Medical Sciences Grant GM65859.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Chemical Communication in a Post-Genomic World,” held January 17–19, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: AHL, acyl-homoserine lactone autoinducer; AIP, autoinducing peptide; AI-1, 3-hydroxybutanoyl-homoserine lactone; AI-2, 3a-methyl-5,6-dihydrofuro[2,3-d][1,3,2]dioxaborole-2,2,6,6a-tetraol.
References
- 1.Miller, M. B. & Bassler, B. L. (2001) Annu. Rev. Microbiol. 55, 165-199. [DOI] [PubMed] [Google Scholar]
- 2.Bassler, B. L. (1999) Curr. Opin. Microbiol. 2, 582-587. [DOI] [PubMed] [Google Scholar]
- 3.Fuqua, C., Winans, S. C. & Greenberg, E. P. (1996) Annu. Rev. Microbiol. 50, 727-751. [DOI] [PubMed] [Google Scholar]
- 4.Engebrecht, J., Nealson, K. & Silverman, M. (1983) Cell 32, 773-781. [DOI] [PubMed] [Google Scholar]
- 5.Engebrecht, J. & Silverman, M. (1984) Proc. Natl. Acad. Sci. USA 81, 4154-4158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Engebrecht, J. & Silverman, M. (1987) Nucleic Acids Res. 15, 10455-10467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Piper, K. R., Beck von Bodman, S. & Farrand, S. K. (1993) Nature 362, 448-450. [DOI] [PubMed] [Google Scholar]
- 8.Bainton, N. J., Stead, P., Chhabra, S. R., Bycroft, B. W., Salmond, G. P., Stewart, G. S. & Williams, P. (1992) Biochem. J. 288, 997-1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Davies, D. G., Parsek, M. R., Pearson, J. P., Iglewski, B. H., Costerton, J. W. & Greenberg, E. P. (1998) Science 280, 295-298. [DOI] [PubMed] [Google Scholar]
- 10.Passador, L., Cook, J. M., Gambello, M. J., Rust, L. & Iglewski, B. H. (1993) Science 260, 1127-1130. [DOI] [PubMed] [Google Scholar]
- 11.Marketon, M. M., Gronquist, M. R., Eberhard, A. & Gonzalez, J. E. (2002) J. Bacteriol. 184, 5686-5695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.de Kievit, T. R. & Iglewski, B. H. (2000) Infect. Immun. 68, 4839-4849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fuqua, C., Parsek, M. R. & Greenberg, E. P. (2001) Annu. Rev. Genet. 35, 439-468. [DOI] [PubMed] [Google Scholar]
- 14.Parsek, M. R. & Greenberg, E. P. (2000) Proc. Natl. Acad. Sci. USA 97, 8789-8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lazazzera, B. A. & Grossman, A. D. (1998) Trends Microbiol. 6, 288-294. [DOI] [PubMed] [Google Scholar]
- 16.Kleerebezem, M., Quadri, L. E., Kuipers, O. P. & de Vos, W. M. (1997) Mol. Microbiol. 24, 895-904. [DOI] [PubMed] [Google Scholar]
- 17.Sturme, M. H., Kleerebezem, M., Nakayama, J., Akkermans, A. D., Vaugha, E. E. & de Vos, W. M. (2002) Antonie Leeuwenhoek 81, 233-243. [DOI] [PubMed] [Google Scholar]
- 18.Magnuson, R., Solomon, J. & Grossman, A. D. (1994) Cell 77, 207-216. [DOI] [PubMed] [Google Scholar]
- 19.Solomon, J. M., Magnuson, R., Srivastava, A. & Grossman, A. D. (1995) Genes Dev. 9, 547-558. [DOI] [PubMed] [Google Scholar]
- 20.Haverstein, L. S. & Morrison, D. A. (1999) in Cell-Cell Signaling in Bacteria, eds. Dunney, G. M. & Winans, S. C. (Am. Soc. Microbiol., Washington, DC), pp. 9-26.
- 21.Novick, R. P. (1999) in Cell-Cell Signaling in Bacteria, eds. Dunney, G. M. & Winans, S. C. (Am. Soc. Microbiol., Washington, DC), pp. 129-146.
- 22.Qin, X., Singh, K. V., Weinstock, G. M. & Murray, B. E. (2000) Infect. Immun. 68, 2579-2586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hanzelka, B. L. & Greenberg, E. P. (1996) J. Bacteriol. 178, 5291-5294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.More, M. I., Finger, L. D., Stryker, J. L., Fuqua, C., Eberhard, A. & Winans, S. C. (1996) Science 272, 1655-1658. [DOI] [PubMed] [Google Scholar]
- 25.Val, D. L. & Cronan, J. E., Jr. (1998) J. Bacteriol. 180, 2644-2651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Watson, W. T., Minogue, T. D., Val, D. L., von Bodman, S. B. & Churchill, M. E. (2002) Mol. Cell 9, 685-694. [DOI] [PubMed] [Google Scholar]
- 27.Zhang, R. G., Pappas, T., Brace, J. L., Miller, P. C., Oulmassov, T., Molyneaux, J. M., Anderson, J. C., Bashkin, J. K., Winans, S. C. & Joachimiak, A. (2002) Nature 417, 971-974. [DOI] [PubMed] [Google Scholar]
- 28.Devine, J. H., Shadel, G. S. & Baldwin, T. O. (1989) Proc. Natl. Acad. Sci. USA 86, 5688-5692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gray, K. M., Passador, L., Iglewski, B. H. & Greenberg, E. P. (1994) J. Bacteriol. 176, 3076-3080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lyon, G. J., Mayville, P., Muir, T. W. & Novick, R. P. (2000) Proc. Natl. Acad. Sci. USA 97, 13330-13335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lyon, G. J., Wright, J. S., Christopoulos, A., Novick, R. P. & Muir, T. W. (2002) J. Biol. Chem. 277, 6247-6253. [DOI] [PubMed] [Google Scholar]
- 32.Lyon, G. J., Wright, J. S., Muir, T. W. & Novick, R. P. (2002) Biochemistry 41, 10095-10104. [DOI] [PubMed] [Google Scholar]
- 33.Ji, G., Beavis, R. & Novick, R. P. (1997) Science 276, 2027-2030. [DOI] [PubMed] [Google Scholar]
- 34.Jarraud, S., Lyon, G. J., Figueiredo, A. M., Gerard, L., Vandenesch, F., Etienne, J., Muir, T. W. & Novick, R. P. (2000) J. Bacteriol. 182, 6517-6522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mayville, P., Ji, G., Beavis, R., Yang, H., Goger, M., Novick, R. P. & Muir, T. W. (1999) Proc. Natl. Acad. Sci. USA 96, 1218-1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brint, J. M. & Ohman, D. E. (1995) J. Bacteriol. 177, 7155-7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pesci, E. C. & Iglewski, B. H. (1997) Trends Microbiol. 5, 132-134; discussion, 134-135. [DOI] [PubMed] [Google Scholar]
- 38.Smith, R. S., Fedyk, E. R., Springer, T. A., Mukaida, N., Iglewski, B. H. & Phipps, R. P. (2001) J. Immunol. 167, 366-374. [DOI] [PubMed] [Google Scholar]
- 39.Miller, M. B., Skorupski, K., Lenz, D. H., Taylor, R. K. & Bassler, B. L. (2002) Cell 110, 303-314. [DOI] [PubMed] [Google Scholar]
- 40.Cao, J. G. & Meighen, E. A. (1989) J. Biol. Chem. 264, 21670-21676. [PubMed] [Google Scholar]
- 41.Chen, X., Schauder, S., Potier, N., Van Dorsselaer, A., Pelczer, I., Bassler, B. L. & Hughson, F. M. (2002) Nature 415, 545-549. [DOI] [PubMed] [Google Scholar]
- 42.Surette, M. G., Miller, M. B. & Bassler, B. L. (1999) Proc. Natl. Acad. Sci. USA 96, 1639-1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bassler, B. L., Wright, M., Showalter, R. E. & Silverman, M. R. (1993) Mol. Microbiol. 9, 773-786. [DOI] [PubMed] [Google Scholar]
- 44.Bassler, B. L., Wright, M. & Silverman, M. R. (1994) Mol. Microbiol. 13, 273-286. [DOI] [PubMed] [Google Scholar]
- 45.Freeman, J. A. & Bassler, B. L. (1999) J. Bacteriol. 181, 899-906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Freeman, J. A., Lilley, B. N. & Bassler, B. L. (2000) Mol. Microbiol. 35, 139-149. [DOI] [PubMed] [Google Scholar]
- 47.Freeman, J. A. & Bassler, B. L. (1999) Mol. Microbiol. 31, 665-677. [DOI] [PubMed] [Google Scholar]
- 48.Lilley, B. N. & Bassler, B. L. (2000) Mol. Microbiol. 36, 940-954. [DOI] [PubMed] [Google Scholar]
- 49.Bassler, B. L., Greenberg, E. P. & Stevens, A. M. (1997) J. Bacteriol. 179, 4043-4045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mok, K. C., Wingreen, N. S. & Bassler, B. L. (2003) EMBO J. 22, 870-881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xavier, K. B. & Bassler, B. L. (2003) Curr. Opin. Microbiol. 6, 191-197. [DOI] [PubMed] [Google Scholar]
- 52.Taga, M. E., Semmelhack, J. L. & Bassler, B. L. (2001) Mol. Microbiol. 42, 777-793. [DOI] [PubMed] [Google Scholar]
- 53.Eberl, L., Winson, M. K., Sternberg, C., Stewart, G. S., Christiansen, G., Chhabra, S. R., Bycroft, B., Williams, P., Molin, S. & Givskov, M. (1996) Mol. Microbiol. 20, 127-136. [DOI] [PubMed] [Google Scholar]
- 54.Givskov, M., de Nys, R., Manefield, M., Gram, L., Maximilien, R., Eberl, L., Molin, S., Steinberg, P. D. & Kjelleberg, S. (1996) J. Bacteriol. 178, 6618-6622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Givskov, M., Eberl, L. & Molin, S. (1997) FEMS Microbiol. Lett. 148, 115-122. [Google Scholar]
- 56.Manefield, M., de Nys, R., Kumar, N., Read, R., Givskov, M., Steinberg, P. & Kjelleberg, S. (1999) Microbiology 145, 283-291. [DOI] [PubMed] [Google Scholar]
- 57.Rasmussen, T. B., Manefield, M., Andersen, J. B., Eberl, L., Anthoni, U., Christophersen, C., Steinberg, P., Kjelleberg, S. & Givskov, M. (2000) Microbiology 146, 3237-3244. [DOI] [PubMed] [Google Scholar]
- 58.Borchardt, S. A., Allain, E. J., Michels, J. J., Stearns, G. W., Kelly, R. F. & McCoy, W. F. (2001) Appl. Environ. Microbiol. 67, 3174-3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dong, Y. H., Gusti, A. R., Zhang, Q., Xu, J. L. & Zhang, L. H. (2002) Appl. Environ. Microbiol. 68, 1754-1759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Dong, Y. H., Wang, L. H., Xu, J. L., Zhang, H. B., Zhang, X. F. & Zhang, L. H. (2001) Nature 411, 813-817. [DOI] [PubMed] [Google Scholar]
- 61.Dong, Y. H., Xu, J. L., Li, X. Z. & Zhang, L. H. (2000) Proc. Natl. Acad. Sci. USA 97, 3526-3531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lee, S. J., Park, S. Y., Lee, J. J., Yum, D. Y., Koo, B. T. & Lee, J. K. (2002) Appl. Environ. Microbiol. 68, 3919-3924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lin, Y. H., Xu, J. L., Hu, J., Wang, L. H., Ong, S. L., Leadbetter, J. R. & Zhang, L. H. (2003) Mol. Microbiol. 47, 849-860. [DOI] [PubMed] [Google Scholar]
- 64.Leadbetter, J. R. & Greenberg, E. P. (2000) J. Bacteriol. 182, 6921-6926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Riedel, K., Hentzer, M., Geisenberger, O., Huber, B., Steidle, A., Wu, H., Hoiby, N., Givskov, M., Molin, S. & Eberl, L. (2001) Microbiology 147, 3249-3262. [DOI] [PubMed] [Google Scholar]