Abstract
In this article we briefly review theories about the ecological roles of microbial secondary metabolites and discuss the prevalence of multiple secondary metabolite production by strains of Streptomyces, highlighting results from analysis of the recently sequenced Streptomyces coelicolor and Streptomyces avermitilis genomes. We address this question: Why is multiple secondary metabolite production in Streptomyces species so commonplace? We argue that synergy or contingency in the action of individual metabolites against biological competitors may, in some cases, be a powerful driving force for the evolution of multiple secondary metabolite production. This argument is illustrated with examples of the coproduction of synergistically acting antibiotics and contingently acting siderophores: two well-known classes of secondary metabolite. We focus, in particular, on the coproduction of β-lactam antibiotics and β-lactamase inhibitors, the coproduction of type A and type B streptogramins, and the coregulated production and independent uptake of structurally distinct siderophores by species of Streptomyces. Possible mechanisms for the evolution of multiple synergistic and contingent metabolite production in Streptomyces species are discussed. It is concluded that the production by Streptomyces species of two or more secondary metabolites that act synergistically or contingently against biological competitors may be far more common than has previously been recognized, and that synergy and contingency may be common driving forces for the evolution of multiple secondary metabolite production by these sessile saprophytes.
Since the discovery of actinomycin in Selman Waksman's laboratory at Rutgers University in 1940, followed in 1943 by streptomycin, the first really effective drug to treat tuberculosis, the actinomycetes have been famous as producers of antibiotics and other “secondary metabolites” with biological activity. During the Golden Age of antibiotic discovery, in the 1950s and 1960s, such well-known antibacterial drugs as tetracycline, erythromycin, and kanamycin, antifungal agents like candicidin and nystatin, and anticancer drugs such as adriamycin were discovered through the efforts of academic and industrial researchers. After 1970 the rate of discovery of useful compounds declined progressively, although several important agents nevertheless came to light, including the antihelmintic avermectin, the immunosuppressants rapamycin and tacrolimus (FK506), and the natural herbicide bialaphos. In fact, the total number of known biologically active molecules continued to grow steadily after the end of the Golden Age. By the mid to late 1990s, thousands of antibiotics and compounds with other biological activities had been described. Estimates of the numbers vary. For example, Demain and Fang (1) gave the total number of antibiotics produced by bacteria and fungi as 5,000, whereas Berdy (2) had more than twice that number. Nevertheless there is agreement that the actinomycetes are responsible for more than two-thirds of the total. Within the actinomycetes, members of the genus Streptomyces account for 70–80% of secondary metabolites, with smaller contributions from genera such as Saccharopolyspora, Amycolatopsis, Micromonospora, and Actinoplanes. What are secondary metabolites, what is their evolutionary significance, and why should the actinomycetes, those soil-dwelling, sporulating members of the high G + C branch of the Gram-positive bacteria, be such prolific producers of them?
There is no pithy one-line definition of the term secondary metabolite, but it nevertheless remains an indispensable epithet in discussions about microbial (and plant) biochemistry and ecology. It embraces the ideas that such compounds are characteristic of narrow taxonomic groups of organisms, such as strains within species, and have diverse, unusual, and often complex chemical structures. They are nonessential for growth of the producing organism, at least under the conditions studied, and are indeed typically made after the phase of most active vegetative growth when the producer is entering a dormant or reproductive stage (1). Their range of biological activities is wide, including the inhibition or killing of other microorganisms (the narrow definition of an antibiotic), but also toxic effects against multicellular organisms like invertebrates and plants. Then there are hormone-like roles in microbial differentiation, and roles in metal transport, a function that blurs the distinction between primary and secondary metabolism. More problematic are the many compounds that either have (so far) no demonstrated biological activity or an activity that is hard to relate to any competitive advantage to the producer, such as a specific effect on the vertebrate immune system exerted by a compound made by a saprophytic soil inhabitant.
The last category was in particular responsible for a widespread view that secondary metabolites were either neutral in evolutionary terms or significant merely as waste products or to keep metabolism ticking over. The implausibility of such general explanations was admirably discussed by Williams et al. (3), who emphasized two powerful arguments for the adaptive significance of secondary metabolites: the complex genetic determination of their biosynthesis, and the exquisite adaptation of many classes of compounds (six were described in detail) to interact with their targets. The former point has been further reinforced by innumerable genetic studies of the biosynthesis of natural products since 1989. Take, for example, erythromycin (4). The producer, Saccharopolyspora erythrea, devotes some 60 kb of its DNA to making this macrolide from propionate units by an amazing assembly-line process involving no fewer than 28 active sites arranged along three giant proteins, followed by hydroxylation and glycosylation involving 18 further proteins, and then has to protect its ribosomes from the highly specific toxicity of the antibiotic by an equally specific methylation of a site on the ribosomal RNA. This is not merely a means of dealing with any excess propionate the cell may produce, nor an idling of the metabolic machinery!
Firn and Jones (5) have made an important recent contribution to the debate. They point out that, whereas some secondary metabolites are indeed highly potent at the concentrations produced in nature and, in the case of antibiotics, against target organisms that actually interact with the producer in the wild, many have only moderate activity. They proposed a unifying model in which it is acknowledged that high-affinity, reversible, noncovalent interactions between a ligand and a protein only occur when the ligand has exactly the right molecular configuration to interact with the complex 3D structure of the protein (what they call “biomolecular activity”) and that this is a rare property. On the other hand, many more molecules have biological activity when tested against a whole organism containing thousands of potential targets that might be inhibited relatively inefficiently. There will therefore be a selective advantage in the evolution of traits that optimize the production and retention of chemical diversity at minimal cost, to allow for the gradual emergence of true biomolecular activity. As long as we accept a selective advantage for the ability to produce secondary metabolites, then, it is legitimate to ask why the streptomycetes and their relatives are preeminent in this ability. The answer almost certainly reflects both the habitat of the organisms and their lifestyle.
The classical habitat of Streptomyces species is as free-living saprophytes in terrestrial soils. Although this is doubtless correct, there is now abundant evidence that some species colonize the rhizosphere of plant roots and even plant tissues; in some cases antibiotic production by the streptomycete may protect the host plant against potential pathogens; the symbiont in turn acquires nutrients from the plant (6, 7). There is now good evidence also for the growth of actinomycetes in marine soils (8).
The soil is a proverbially complex environment in which innumerable stresses (chemical, physical, and biological) occur in a temporally and spatially variable manner. Moreover, streptomycetes are nonmotile, so stresses cannot be avoided but have to be met. The need to combat stress was the explanation invoked by Bentley et al. (9) for the enormous numbers of genes that would encode regulators, transport proteins, and nutritional enzymes identified in the Streptomyces coelicolor genome sequence. A striking feature of this 8.7-megabase (Mb) genome is its notional division into a “core” region of ≈4.9 Mb and left and right “arms” of ≈1.5 and 2.3 Mb, respectively. Classes of genes that would encode unconditionally essential functions such as the machinery of DNA replication, transcription, and translation were found in the core, whereas examples of conditionally adaptive functions, such as the ability to grow on complex carbohydrates like cellulose, chitin, and xylan, occurred predominately in the arms. The genome sequence also revealed ≈23 clusters of genes, representing ≈4.5% of the genome, that were predicted to encode biosynthetic enzymes for a wide range of secondary metabolites (Table 1), only half a dozen of which had previously been identified. Interestingly in the present context, many of the clusters reside in the arms or close to their boundary with the core of the genome, as pointed out by Piepersberg (10) in a review that emphasizes the roles of secondary metabolites in chemical communication, the topic of this Sackler symposium. An even larger number of secondary metabolic gene clusters was found in the recently sequenced Streptomyces avermitilis genome (30 clusters covering ≈6% of the genome), again many of them in the arm regions (11). Thus, assuming a selective advantage for secondary metabolite production, such an advantage for many of the compounds is likely to be conditional, or sporadic.
Table 1. Gene clusters potentially directing the production of secondary metabolites in S. coelicolor.
| Biosynthetic system | Metabolite | Size, kb | Location |
|---|---|---|---|
| Type II PKS | Actinorhodin | 22 | 5071—5092 |
| Type II PKS | Gray spore pigment | 8 | 5314—5320 |
| Mixed | Methylenomycin | 20 | SCP1 plasmid |
| NRPS; type I modular PKS | Prodiginines | 33 | 5877—5898 |
| NRPS | CDA | 80 | 3210—3249 |
| NRPS | Coelichelin | 20 | 0489—0499 |
| NRPS | Coelibactin | 26 | 7681—7691 |
| NRPS | Unknown | 14 | 6429—6438 |
| Type I modular PKS | Unknown | 70 | 6273—6288 |
| Type I modular PKS | Unknown | 10 | 6826—6827 |
| Type I iterative PKS | Polyunsaturated fatty acid? | 19 | 0124—0129 |
| Chalcone synthase | Tetrahydroxynaphthalene | 1 | 1206—1208 |
| Chalcone synthase | Unknown | 3.5 | 7669—7671 |
| Chalcone synthase | Unknown | 1 | 7222 |
| Sesquiterpene synthase | Geosmin | 2 | 6073 |
| Sesquiterpene synthase | Unknown | 2.5 | 5222—5223 |
| Squalene-Hopene cyclase | Hopanoids | 15 | 6759—6771 |
| Phytoene synthase | Isorenieratine | 0185—0191 | |
| Siderophore synthetase | Desferrioxamines | 5 | 2782—2785 |
| Siderophore synthetase | Unknown | 4 | 5799—5801 |
| Type II fatty acid synthase | Unknown | 10 | 1265—1273 |
| Butyrolactone synthase | Butyrolactones? | 1 | 6266 |
| Deoxysugar | Unknown | 20 | 0381—0401 |
As pointed out by Chater and Merrick (12), Streptomyces antibiotics are typically produced in small amounts at the transition phase in colonial development when the growth of the vegetative mycelium is slowing as a result of nutrient exhaustion and the aerial mycelium is about to develop at the expense of nutrients released by breakdown of the vegetative hyphae (13). Such antibiotics are proposed to defend the food source when other soil microorganisms threaten it. The hypothesis is strengthened by the finding of large numbers of antibiotics in other groups of differentiating microbes such as filamentous fungi and myxobacteria. The concept has been extended to the possibility that competitors might be attracted to the amino acids, sugars, and other small molecules arising from the degraded vegetative mycelium and be killed and recycled by the developing Streptomyces colony, a concept dubbed “fatal attraction” in relation to Myxococcus by Shi and Zusman (14).
Synergy and Contingency§ in Secondary Metabolite Action
Against this background of likely selective advantages, then, actinomycetes have evolved some amazingly potent agents with biomolecular activity and numerous others with a more generalized biological effect. Perhaps more strikingly, there are now numerous examples of the production by an actinomycete of two chemically different metabolites that act either synergistically against a target microorganism, as recently emphasized by McCafferty and coworkers (15), or contingently to overcome competition with other microorganisms for nutrients. In this context, synergistic metabolites have a greater antibiotic activity against competitors in combination than the sum of their individual antibiotic activities (see below), whereas contingently acting metabolites possess similar biological activity (e.g., iron sequestration as below), but are independently recognized and used by the producing organism and competitors in its environment. In the rest of this article we discuss some examples of these phenomena and attempt to relate them to the ideas about the roles and evolution of secondary metabolism outlined above.
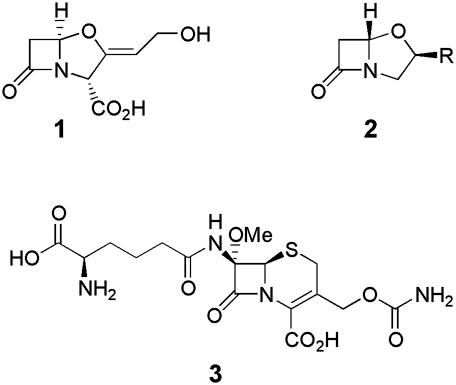
β-Lactam Antibiotics and Clavulanic Acid. Clavulanic acid (1) (Fig. 1) is a natural inhibitor of β-lactamases (enzymes that confer resistance to β-lactam antibiotics in many microorganims). The genetics and biochemistry of its production have been intensively studied in Streptomyces clavuligerus for >25 years (16). This actinomycete also produces several other β-lactam compounds, including a number of structurally related clavams (2) (Fig. 1), which are antipodal to clavulanic acid and are not β-lactamase inhibitors (although they do exhibit antifungal activity), and the cephalosporin antibiotic cephamycin C (3) (Fig. 1) (along with other biologically active intermediates on its biosynthetic pathway), which inhibits the transpeptidation reaction in cell wall biosynthesis. The early steps of clavulanic acid and clavam biosynthesis are common, but the pathways to these two structurally related metabolites diverge at clavaminic acid (16). In contrast, cephamycin is biosynthesized by a pathway that is mechanistically distinct from that for the clavams and clavulanic acid (16, 17). Combinations of β-lactam antibiotics and β-lactamase inhibitors are well known to be effective against β-lactam-resistant bacteria in comparison with β-lactams antibiotics alone. This is because of the synergistic action of these metabolites, which is reflected by the name chosen for the clinically used combination of clavulanic acid with methicillin: augmentin.
Fig. 1.
Structures of clavulanic acid (1), clavams (R = variable group) (2), and cephamycin C (3), coproduced by several Streptomyces species. 1 and 3 act synergistically to inhibit cell wall biosynthesis in β-lactam-resistant bacteria.
At first sight the coproduction of cephamycin C and clavulanic acid by S. clavuligerus might seem like an unusual coincidence. Closer inspection of metabolite production patterns among other producers of clavulanic acid, clavams, and cephamycin C, however, suggests that a strong selective pressure, rather than mere chance, has created actinomycetes that coproduce clavulanic acid and a β-lactam antibiotic such as cephamycin C. Thus, S. clavuligerus, Streptomyces jumonjinensis, Streptomyces katsurahamanus, and an unclassified Streptomyces sp. all produce clavulanic acid (16). Strikingly, all of these streptomycetes also produce cephamycin C (16). Indeed there are no known producers of clavulanic acid that do not also produce cephamycins (16). In contrast, several Streptomyces species produce clavams, but not clavulanic acid or cephamycins (16), and some actinomycetes, such as Nocardia lactamdurans and Streptomyces griseus NRRL 3851, produce cephamycin C but not clavams or clavulanic acid (17). The fact that no known actinomycetes produce clavulanic acid alone, but there are actinomycetes that produce just cephamycin C or clavams, suggests that the production of clavulanic acid evolved in an ancestral clavam and cephamycin producer as a response to the acquisition of β-lactamase-mediated resistance in bacteria inhabiting the same environmental niche and thus posing biological competition. The existence of several clavam-only-producing strains suggests that one of these may initially have acquired the cephamycin pathway by horizontal transfer, which conferred a selective advantage against β-lactam-sensitive bacteria. These sensitive bacteria then acquired β-lactamase-mediated resistance to cephamycin, and eventually the production of clavulanic acid by a modification of the clavam biosynthetic pathway in the clavam/cephamycin producer was selected for by biological competition from the β-lactam-resistant bacteria. The production of clavulanic acid by the cephamycin/clavam producer would restore the effectiveness of cephamycin as an antibiotic against the β-lactam-resistant competition.
Support for the above hypothesis for the evolution of clavulanic acid production derives from analysis of the genes that direct clavam and clavulanic acid production. Thus, the clavulanic acid gene cluster is directly adjacent to the cephamycin cluster on the chromosomes of S. clavuligerus, S. jumonjinensis, and S. katsurahamanus (18). Such “superclustering” would be expected for gene clusters that direct the production of metabolites that act synergistically to benefit the producing organism (19, 20). In addition, the production of both cephamycin and clavulanic acid in S. clavuligerus is controlled principally by the ccaR (dclX) gene, which codes for an OmpR-like transcriptional regulator, and is located within the cephamycin cluster (21, 22). In contrast, the clavam cluster is at least 20–30 kb away from the cephamycin/clavulanic acid clusters on the S. clavuligerus chromosome, and the regulation of clavam production is distinct from the coregulated production of cephamycins and clavulanic acid (23–25). These observations suggest that clavulanic acid production in a clavam-producing ancestor, which acquired the ability to produce cephamycin, could have arisen via chromosomal duplication of the clavam cluster followed by subsequent acquisition of the late steps in the clavulanic acid pathway, which involve the stereochemical inversions that are key to β-lactamase activity. The chromosomal linkage of the cephamycin and the clavulanic acid clusters would facilitate simultaneous horizontal transfer of both clusters to other organisms, which would clearly be beneficial to recipients. Also, there would be pressure to evolve coregulation of clavulanic acid and cephamycin production, as the coordinated production of a β-lactam antibiotic and a β-lactamase inhibitor is clearly beneficial for survival. Thus, there appears to be clear selective pressure and a plausible genetic mechanism for the evolution of the coproduction of clavulanic acid and cephamycin in Streptomyces species.
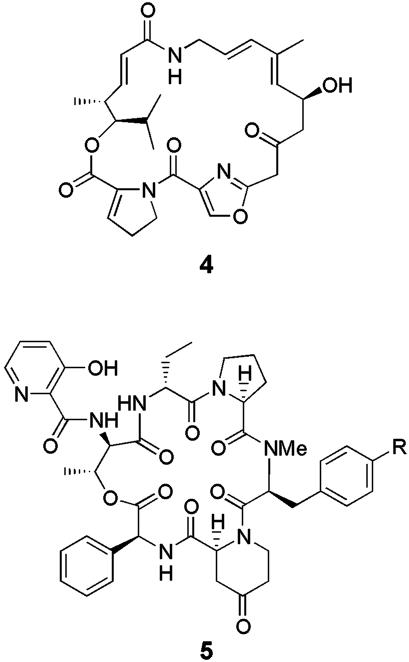
Streptogramins. Streptogramins are pairs of structurally unrelated antibiotics that synergistically inhibit bacterial ribosomal protein synthesis at the peptidyl transfer step (15). The type A streptogramins [e.g., pristinamycin IIA/virginiamycin M1 (4) (Fig. 2)] are assembled through a mixed polyketide/nonribosomal peptide pathway, whereas the type B streptogramins [e.g., pristinamycin IA (5) (Fig. 2) (R = NMe2) and virginiamycin S1 (6) (R = H)] are nonribosomally synthesized depsipeptides that contain several unusual nonproteinogenic amino acids (26–33). 4 is coproduced with 5 by Streptomyces pristinaespiralis NRRL 2958 and with 6 by Streptomyces virginiae (34). Several other streptomycetes, such as Streptomyces mitakaensis, Streptomyces graminofaciens, and Streptomyces loidensis, have been reported to coproduce similar type A and type B streptogramins. No species are known that produce only a type A or a type B streptogramin. Whereas type A or type B streptogramins alone are bacteriostatic, the combination of a type A and type B streptogramin is bacteriocidal. This increase in antibacterial efficacy has been shown to originate from synergistic binding of the type A and type B streptogramins to distinct sites on the ribosome (35, 36). Thus, binding of streptogramin A antibiotics to the ribosome increases the affinity of streptogramin B antibiotics for the ribosome by up to 40-fold, rendering dual antibiotic binding essentially irreversible (37). The binding of the type A antibiotics is thought to change the conformation of the ribosomal RNA to expose a high-affinity binding site for the type B compounds (37). The synergistic antibiotic effect of the streptogramins has been exploited clinically in the form of a combination of semisynthetic pristinamycin IA and IIA derivatives known as synercid, which is used for the treatment of multidrug-resistant bacterial infections.
Fig. 2.
Structures of pristinamycin I (4) and II (R = variable group) (5) components, which inhibit protein synthesis in bacteria by binding synergistically to the ribosome.
Evidence to support the idea that coproduction of streptogramin A and B antibiotics in several bacteria has evolved under strong selective pressure stems from analysis of the chromosomal arrangement of genes that direct streptogramin production. Thus, Bamas–Jacques et al. (38) demonstrated that genes directing the production of pristinamycins I and II are clustered in a 200-kb region of the chromosome of S. pristinaespiralis. The finding that genes for the biosynthesis of the pristinamycin I and II components are interspersed in this cluster led Bamas-Jacques et al. to suggest that the pathways to the type A and type B streptogramins coevolved in the same organism, perhaps from a common origin. The structural dissimilarity of the type A and type B streptogramins, however, implies that they are biosynthesized by quite different catalytic machinery. Indeed analysis of the genes that direct production of the pristinamycin type I and type II components indicates that they are biosynthesized by nonribosomal peptide and predominantly polyketide pathways, respectively (30–33). It is therefore hard to imagine how the pathways to the type I and type II metabolites could have evolved from a common origin. An alternative, stepwise model for the acquisition of the type A and type B streptogramin pathways cannot be ruled out on the basis of available evidence. Thus, an ancestral streptomycete may have originally produced only one of the streptogramin components, which was initially effective at inhibiting ribosomal protein synthesis in competing organisms. Over time the competing organisms would acquire resistance to the single streptogramin component, rendering it less effective. The pathway to the second streptogramin component could then have been acquired by chance horizontal transfer. The synergisitic effect of the pair of streptogramins would give the recipient a renewed advantage over competing organisms, which would lead to retention of both pathways. Initially, the two pathways may have been located in distinct regions of the chromosomes. Subsequent chromosomal rearrangements (which are well known in Streptomyces species) could intersperse the two clusters with each other, which could facilitate the coregulated production of both streptogramin components and the horizontal transfer of both pathways into other organisms. The stepwise model for evolution of streptogramin production is similar to that outlined above for evolution of cephamycin and clavulanic acid production and might prove to be a useful general model for evolution of the production of multiple secondary metabolites that function synergistically.
Other Potentially Synergistic, Coproduced Pairs or Groups of Antibiotics. Streptomycetes are famed for producing multiple antibiotics, and it seems likely that there are many other examples of individual Streptomyces species that produce two or more antibiotics that act synergistically against a competing organism. One interesting potential example has arisen from sequencing of the Streptomyces avermitilis genome (11). Analysis of gene clusters that code for polyketide synthase (PKS) multienzyme systems, commonly associated with antibiotic biosynthesis, revealed that S. avermitilis has the capability to produce two structurally distinct antifungal compounds: oligomycin and a polyene macrolide (11). These antibiotics act on distinct molecular targets in eukaryotes. Thus, oligomycin inhibits mitochondrial F0F1-ATP synthase, whereas polyene macrolides bind irreversibly to fungal cell membranes, altering their permeability (39, 40). It seems possible that oligomycin and polyene macrolides could act synergistically against fungi and that a persistent fungal competitor in the natural environment of S. avermitilis has selected for the coproduction of these compounds. It is note-worthy, however, that the gene clusters directing oligomycin and polyene macrolide biosynthesis are separated by >2,500 kb on the S. avermitilis chromosome, and it will be interesting to see whether the production of these two antifungal compounds is coregulated (41).
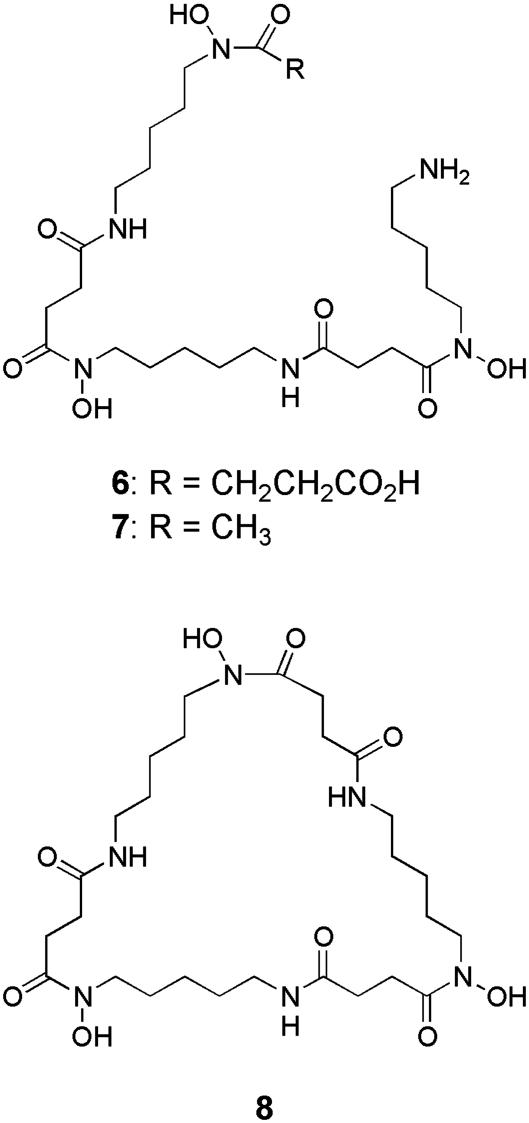
Production of Multiple Siderophores. Siderophores are small diffusible molecules excreted by many microorganisms that form very stable complexes with ferric iron. In Gram-positive bacteria, the iron–siderophore complexes are selectively recognized by membrane-associated receptors and actively transported into the cell by ATP-dependent transmembrane transporters (42). Once inside the cell, the iron–siderophore complex is dissociated, often by hydrolysis of the multidentate siderophore ligand and/or reduction of ferric iron to ferrous iron, which is stored in bacterioferritin and used as a cofactor in several vital cellular processes (43). Siderophores are produced by saprophytes to overcome the inherent aqueous insolubility of ferric iron, which limits its availability in soil. More than 10 distinct species of Streptomyces have been reported to produce characteristic desferrioxamine siderophores such as desferrioxamines G1 (6), B (7), and E (8) (44) (Fig.3). Until very recently, however, siderophores belonging to other structural classes such as peptide hydroxamates, catecholates, α-hydroxycarboxylates, and thiazolines/oxazolines have not been reported as metabolic products of Streptomyces species.
Fig. 3.
Structures of desferrioxamine siderophores typically produced by Streptomyces species.
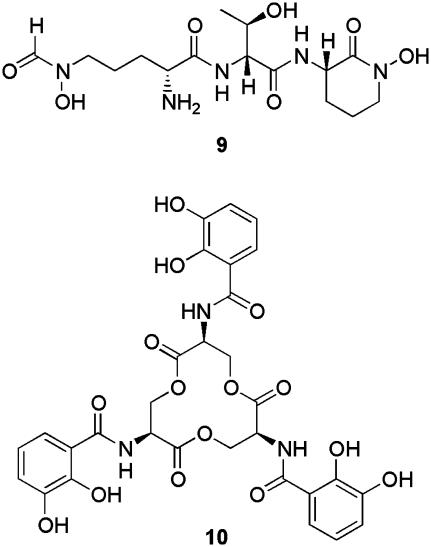
In silico analysis of the S. coelicolor genome sequence suggested that, in addition to a pathway for desferrioxamine biosynthesis, multiple nonribosomal peptide synthetase (NRPS) pathways for the assembly of structurally diverse siderophores exist (9). One of these pathways has been predicted to culminate in the peptide hydroxamate siderophore coelichelin (9) (Fig. 4) (45). Recently, Challis and coworkers have confirmed, using a combination of gene inactivation and metabolic profiling experiments, that two independent pathways exist in S. coelicolor for the production of hydroxamate siderophores (S. Lautru, F. Barona-Gomez, U. Wong, and G.L.C., unpublished work). Thus, inactivation of desD, which codes for a siderophore synthetase believed to catalyze the key step in desferrioxamine biosynthesis, causes abrogation of the production of 6 and 8, whereas inactivation of cchH, which codes for the NRPS predicted to assemble 9, leads to loss of production of a different hydroxamate siderophore (F. Barona-Gomez, U. Wong, A. Giannakupolous, P. J. Derrick, and G.L.C., unpublished work).
Fig. 4.
Predicted structure of coelichelin (9) and structure of enterobactin (10), siderophores of diverse structure coproduced with desferrioxamines by some Streptomyces species that are thought to provide a contingency plan for iron uptake in the event of desferrioxamine piracy.
The production of both the desferrioxamines and the other hydroxamate siderophore is maximal under iron-deficient conditions and completely suppressed under iron-sufficient conditions. Consistent with this observation, analysis of the cch and des clusters indicates that, although they are not closely linked on the S. coelicolor chromosome, their transcription is very likely to be coregulated, because intergenic regions in both clusters contain similar inverted repeat sequences that match the consensus sequence 5′-TTAGGTTAGGCTCACCTAA-3′ for iron-dependent repressor (IdeR) binding. IdeR is known to regulate the transcription of siderophore biosynthesis genes in other Gram-positive bacteria, e.g., Mycobacterium species (46).
It is not immediately obvious what selective advantage S. coelicolor gains through the coregulated production of two structurally distinct hydroxamate siderophores. Yet this is not an isolated example of multiple siderophore production by Streptomyces species. Recently, Fiedler and coworkers (47) reported that Streptomyces tendae Tü 901/8c and Streptomyces sp. Tü 6125 produce enterobactin 10 (the characteristic siderophore of Enterobacteriaceae; Fig. 4) in addition to the characteristic Streptomyces siderophores 7 and 8. It seems probable that enterobactin is biosynthesized in streptomycetes via a NRPS pathway, similar to the well-characterized Escherichia coli pathway, and that this pathway is distinct from that for desferrioxamine biosynthesis (48). A third potential example of multiple siderophore production by Streptomyces species has been uncovered through preliminary examination of the S. avermitilis genome sequence, which, in addition to a cluster of genes virtually identical to the des cluster of S. coelicolor, contains a gene cluster encoding a NRPS system predicted to produce a siderophore of similar structure to the myxobacterial siderophore myxochelin (11, 49). Thus, the production of structurally diverse secondary nonribosomally synthesized peptide siderophores, in addition to the characteristic desferrioxamine siderophores, may be common in streptomycetes.
An appealing explanation for the coregulated production of two or more structurally distinct siderophores by Streptomyces species stems from the observation that many organisms that neither biosynthesise nor excrete desferrioxmine-like siderophores are nevertheless able to specifically take up ferrioxamine complexes and use the iron associated with them. These organisms would pose a serious biological challenge to sessile streptomycetes that possess only a desferrioxamine pathway for scavenging iron from the environment. This would exert strong selective pressure on such streptomycetes for acquisition of a second cluster of genes directing the production of another siderophore (e.g., by horizontal transfer), whose ferric complex could be selectively recognized and taken up by the cell through a separate transport system from that for ferrioxamine uptake. Consistent with this model, analysis of the des and cch clusters of S. coelicolor reveals that a gene coding for a distinct iron–siderophore-binding lipoprotein is present in each cluster. Iron–siderophore-binding lipoproteins are receptors associated, via a covalently attached lipid, with the extracellular membrane of Gram-positive bacteria that selectively recognize iron–siderophore complexes and initiate their ATP-driven transport into the cell. Thus, the finding that iron–siderophore-binding lipoproteins are coded for in both the des and cch clusters suggests that the ferric complexes of each of these siderophores can be selectively and independently taken up by S. coelicolor. The biological competition from other organisms able to take up ferrioxamines would be lessened by coregulated production of coelichelin and desferrioxamines, because the ferric coelichelin complex can be selectively absorbed into S. coelicolor cells through an independent uptake pathway. In this scenario, coelichelin and desferrioxamine are acting contingently rather than synergistically, because they allow S. coelicolor to survive in environments inhabited by unforeseen competitors whose ability to use ferrioxamine (or indeed other ferric siderophore complexes) cannot be readily anticipated.
The notion that a second cluster of genes directing siderophore production in primordial desferrioxamine-producing streptomycetes was acquired by horizontal transfer is supported by comparison of the structures of enterobactin, and that predicted for coelichelin (produced by S. tendae and S. coelicolor, respectively). Whereas enterobactin is the characteristic siderophore of E. coli, the predicted structure of coelichelin is similar to several hydroxamate siderophores produced by mycobacteria, suggesting that the “second” siderophores of streptomycetes derive from diverse origins. It will be interesting to see whether other, structurally diverse siderophores are isolated, along with the characteristic desferrioxamines, from further Streptomyces species in the future.
Conclusions
The well known property of many Streptomyces species to produce multiple antibiotics or other secondary metabolites has attracted much recent attention, not least because analysis of the recently completed S. coelicolor and S. avermitilis genome sequences has suggested that this ability may be far greater than was previously thought. Clear evidence emerging from the recent literature suggests that two driving forces for the evolution of this phenomenon may be synergistic and contingent action against biological competitors, which Streptomyces species are not easily able to evade in their saprophytic lifestyle because of a lack of motility. Indeed, as the study of Streptomyces secondary metabolism continues, it is anticipated that many more cases of two or more structurally distinct metabolites that act synergistically or contingently against biological competition will be discovered.
Acknowledgments
Francisco Barona-Gomez and Sylvie Lautru are gratefully acknowledged for helpful discussions and suggestions on the manuscript.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Chemical Communication in a Post-Genomic World,” held January 17–19, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: PKS, polyketide synthase; NRPS, nonribosomal peptide synthetase; IdeR, iron-dependent repressor.
Footnotes
Note that our use of “contingency” in this article relates to multiple metabolites acting on the same biological target to provide an organism with a contingency plan to combat unforeseeable biological competition. Moxon and coworkers (50) have used contingency to describe hypermutable loci coding for variable surface proteins in Haemophilus influenzae and Nesseria meningitidis. The two uses of the word should not be confused.
References
- 1.Demain, A. & Fang, A. (2000) in History of Modern Biotechnology, ed. Fichter, A. (Springer, Berlin), Vol. 1, pp. 2-39. [Google Scholar]
- 2.Berdy, J. (1995) in Proceedings of the Ninth International Symposium on the Biology of the Actinomycetes, eds. Debabov, V. G., Dudnik, Y. V. & Danilenko, V. N. (All-Russia Research Institute for Genetics and Selection of Industrial Microrganisms, Moscow), pp. 13-34.
- 3.Williams, D. H., Stone, M. J., Hauck, P. R. & Rahman, S. K. (1989) J. Nat. Products 52, 1189-1208. [DOI] [PubMed] [Google Scholar]
- 4.Donadio, S., Staver, M. J., McAlpine, J. B., Swanson, S. J. & Katz, L. (1991) Proc. Natl. Acad. Sci. USA 252, 675-679. [DOI] [PubMed] [Google Scholar]
- 5.Firn, R. D. & Jones, C. G. (2000) Mol. Microbiol. 37, 989-994. [DOI] [PubMed] [Google Scholar]
- 6.Tokala, R. K., Strap, J. L., Jung, C. M., Crawford, D. L., Salove, M. H., Deobald, L. A., Bailey, J. F. & Morra, M. J. (2002) Appl. Environ. Microbiol. 68, 2161-2171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Castillo, U. F., Strobel, G. A., Ford, E. J., Hess, W. M., Porter, H., Jensen, J. B., Albert, H., Robison, R., Condron, M. A. M., Teplow, D. B., et al. (2002) Microbiology 148, 2675-2685. [DOI] [PubMed] [Google Scholar]
- 8.Mincer, T. J., Jensen, P. R., Kauffman, C. A. & Fenical, W. (2002) Appl. Env. Microbiol. 68, 5005-5011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bentley, S. D., Chater, K. F., Cerdeno-Tarraga, A.-M., Challis, G. L., Thomson, N. R., James, K. D., Harris, D. E., Quail, M. A., Kieser, H., Harper, D., et al. (2002) Nature 417, 141-147. [DOI] [PubMed] [Google Scholar]
- 10.Piepersberg, W. (2002) in Molecular Medical Microbiology, ed. Sussman, M. (Academic, San Diego), Vol. 1, pp. 561-594. [Google Scholar]
- 11.Ikeda, H., Ishikawa, J., Hanamoto, A., Shinose, M., Kikuchi, H., Shiba, T., Sakaki, Y., Hattori, M. & Omura, S. (2003) Nat. Biotechnol. 21, 526-531. [DOI] [PubMed] [Google Scholar]
- 12.Chater, K. F. & Merrick, M. J. (1979) in Developmental Biology of Prokaryotes, ed. Parish, J. H. (Blackwell, Oxford), pp. 93-114.
- 13.Migulez, E. M., Hardisson, C. & Manzanal, M. B. 2000) Int. Microbiol. 3, 153-158. [PubMed] [Google Scholar]
- 14.Shi, W. & Zusman, D. R. (1993) Nature 366, 414-415. [DOI] [PubMed] [Google Scholar]
- 15.McCafferty, D. G., Cudic, P., Yu, Y. K., Behena, D. C. & Kruger, R. (1999) Curr. Opin. Chem. Biol. 3, 672-680. [DOI] [PubMed] [Google Scholar]
- 16.Jensen, S. E. & Paradkar, A. S. (1999) Antonie van Leeuwenhoek 75, 125-133. [DOI] [PubMed] [Google Scholar]
- 17.Liras, P. (1999) Antonie van Leeuwenhoek 75, 109-124. [DOI] [PubMed] [Google Scholar]
- 18.Ward, J. M. & Hodgson, J. E. (1993) FEMS Microbiol. Lett. 110, 239-242. [DOI] [PubMed] [Google Scholar]
- 19.Jensen, S. E., Alexander, D. C., Paradkar, A. S. & Aido, K. A. (1993) in Industrial Microorganisms: Basic and Applied Molecular Genetics, eds. Baltz, R. H., Hegeman, G. D. & Skatrud, P. L. (Am. Soc. Microbiol., Washington, DC), pp. 169-176.
- 20.Lawrence, G. J. (1997) Trends Microbiol. 5, 355-359. [DOI] [PubMed] [Google Scholar]
- 21.Walters, N. J., Barton, B. & Earl, A. J. (1994) International Patent WO94/18326-A1.
- 22.Perez-Llarena, F., Liras, P., Rodriguez-Garcia, A. & Martin, J. F. (1997) J. Bacteriol. 179, 2053-2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mosher, R. H., Paradkar, A. S., Anders, C., Barton, B. & Jensen, S. E. (1999) Antimicrob. Agents Chemother. 43, 1215-1224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Paradkar, A. & Jensen, S. E. (1995) J. Bacteriol. 177, 1307-1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marsh, E. N., Chang, M. D.-T. & Townsend, C. A. 1992) Biochemistry 31, 12648-12657. [DOI] [PubMed] [Google Scholar]
- 26.Blanc, V., Blanche, F., Crouzet, J., Jacques, N., Lacroix, P., Thibaut, D. & Zagorec, M. (1994) International Patent WO 9408014.
- 27.Blanc, V., Gil, P., Bamas-Jacques, N., Lorenzon, S., Zagorec, M., Schleuniger, J., Bisch, D., Blanche, F., Debussche, L., Crouzet, J., et al. (1997) Mol. Microbiol. 23, 191-202. [DOI] [PubMed] [Google Scholar]
- 28.Blanc, V., Lagneaux, D., Didier, P. Gil, P., Lacroix, P. & Crouzet, J. (1995) J. Bacteriol. 177, 5206-5214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanc, V., Thibaut, D., Bamas-Jacques, N., Blanche, F., Crouzet, J., Barriere, J.-C., Debussche, L., Famechon, A., Paris, J.-M., Dutruc-Rosset, G., et al. (1996) International Patent WO 9601901.
- 30.de Crécy-Lagard, V., Blanc, V., Gil, P., Naudin, L., Lorenzon, S., Famechohn, A., Bamas-Jacques, N., Crouzet, J., Thiebaut, D., et al. (1997) J. Bacteriol. 179, 705-713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Crécy-Lagard, V., Saurin, W., Thibaut, D., Gil, P., Naudin, L., Crouzet, J. & Blanc, V. (1997) Antimicrob. Agents Chemother. 41, 1904-2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Thibaut, D., Bisch, D., Ratet, N., Maton, L., Couder, M., Debussche, L. & Blanche, F. (1997) J. Bacteriol. 179, 697-704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thibaut, D. Ratet, N., Bisch, D., Faucher, D., Debussche, L. & Blanche, F. (1995) J. Bacteriol. 177, 5199-5205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cocito, C. G. (1979) Microbiol. Rev. 43, 145-198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Porse, B. T. & Garrett, R. A. (1999) J. Mol. Biol. 286, 375-387. [DOI] [PubMed] [Google Scholar]
- 36.Contreras, A. & Vasquez, D. (1977) Eur. J. Biochem. 74, 549-551. [DOI] [PubMed] [Google Scholar]
- 37.Di Giambattista, M., Chinali, G. & Cocito, C. 1989) J. Antimicrob. Chemother. 24, 485-507. [DOI] [PubMed] [Google Scholar]
- 38.Bamas-Jacques, N., Lorenzon, S., Lacroix, P., de Swetschin, C. & Crouzet, J. (1999) J. Appl. Microbiol. 87, 939-948. [DOI] [PubMed] [Google Scholar]
- 39.Zotchev, S. B. (2003) Curr. Med. Chem. 10, 211-223. [DOI] [PubMed] [Google Scholar]
- 40.Corran, A. J., Renwick, A. & Dunbar, S. J. (1998) Pestic. Sci. 54, 338-344. [Google Scholar]
- 41.Omura, S., Ikeda, H., Ishikawa, J., Hanamoto, A., Takahashi, C., Shinose, M., Takahashi, Y., Horikawa, H., Nakazawa, H., Osonoe, T., et al. (2001) Proc. Natl. Acad. Sci. USA 98, 12215-12220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schneider, R. & Hantke, K. (1993) Mol. Microbiol. 8, 111-121. [DOI] [PubMed] [Google Scholar]
- 43.Winkelman, G. & Drechsel, H. (1997) in Biotechnology, eds. Rehm, H.-J. & Reed, G. (VCH, Weinheim, Germany), 2nd Ed., Vol. 7, pp. 200-246. [Google Scholar]
- 44.Müller, A. & Zähner, H. (1968) Arch. Mikrobiol. 62, 257-263. [PubMed] [Google Scholar]
- 45.Challis, G. L. & Ravel, J. (2001) FEMS Microbiol. Lett. 187, 111-114. [DOI] [PubMed] [Google Scholar]
- 46.Schmitt, M. P., Predich, M., Doukhan, L., Smith, I. & Holmes, R. K. (1995) Infect. Immun. 63, 4284-4289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Fiedler, H.-P., Krastel, P., Muller, J., Gebhardt, K. & Zeeck, A. (2001) FEMS Microbiol. Lett. 196, 147-151. [DOI] [PubMed] [Google Scholar]
- 48.Crosa, J. H. & Walsh, C. T. (2002) Microbiol. Mol. Biol. Rev. 66, 223-249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kunze, B., Bedorf, N., Kohl, W., Hoefle, G. & Reichenbach, H. (1989) J. Antibiot. 42, 14-17. [DOI] [PubMed] [Google Scholar]
- 50.Moxon, E. R., Rainey, P. B., Nowak, M. A. & Lenski, R. (1994) Curr. Biol. 4, 24-33. [DOI] [PubMed] [Google Scholar]