Abstract
The 500 different species of venomous cone snails (genus Conus) use small, highly structured peptides (conotoxins) for interacting with prey, predators, and competitors. These peptides are produced by translating mRNA from many genes belonging to only a few gene superfamilies. Each translation product is processed to yield a great diversity of different mature toxin peptides (≈50,000–100,000), most of which are 12–30 aa in length with two to three disulfide crosslinks. In vitro, forming the biologically relevant disulfide configuration is often problematic, suggesting that in vivo mechanisms for efficiently folding the diversity of conotoxins have been evolved by the cone snails. We demonstrate here that the correct folding of a Conus peptide is facilitated by a posttranslationally modified amino acid, γ-carboxyglutamate. In addition, we show that multiple isoforms of protein disulfide isomerase are major soluble proteins in Conus venom duct extracts. The results provide evidence for the type of adaptations required before cone snails could systematically explore the specialized biochemical world of “microproteins” that other organisms have not been able to systematically access. Almost certainly, additional specialized adaptations for efficient microprotein folding are required.
Chemical interactions between organisms, the major theme of this symposium, are usually mediated through organic molecules that elicit physiological effects in the targeted organisms that are of benefit to the producer. In this respect, a group of predatory molluscs, the cone snails (see Fig. 1), use an idiosyncratic strategy; small structured peptides, instead of conventional natural products, are the primary agents used for interacting with other animals. The cone snails belong to the genus Conus, comprising 500 different species, all of which capture prey by injecting a peptide-rich venom (1). Some, if not most, species of Conus also use venom to defend against predators and for competitive interactions (2). Thus, the “front-line” genes for the biotic interactions of Conus are those encoding their venom peptides. The peptidic nature of the functional gene products means that these can be directly elucidated and chemically synthesized from the gene sequence. Consequently, for the cone snails, the link between chemical ecology and genomics is unusually straightforward.
Fig. 1.
Shells of cone snails. Seven different Conus species (of the ≈500 total) are illustrated: the examples shown represent the three major feeding types of Conus. Experimental data that are described in the text were obtained from these species. (Top) The glory-of-the-sea cone, C. gloriamaris (Left); the cloth-of-gold cone, C. textile (Right). (Middle) C. omaria (Left); C. consors (Center); C. aurisiacus (Right). (Bottom) The fly-speck cone, C. stercusmuscarum (Left); C. betulinus (Right). C. betulinus is worm-hunting, whereas C. consors, C. aurisiacus, and C. stercusmuscarum are piscivorous (fish hunting). C. textile, C. gloriamaris, and C. omaria are molluscivorous (mollusc hunting). A PCR screen of PDI was carried out on most of these species (see text). A comparison of the spasmodic peptide of C. textile and C. gloriamaris provided the basis for the work on the role of posttranslational modification in Conus peptide folding. C. textile was the species used for most of the experimental studies described in this article.
Each Conus species has a distinct set of biotic interactions characteristic of that species; this helps to rationalize why each one has a different repertoire of 100–200 venom peptides (only a subset of which are probably expressed at any one time). Thus, the 500 different living species of cone snails (3) can potentially produce ≈50,000–100,000 different conopeptides in their venom ducts, an enormous diversity of gene products reflecting the complex chemical ecology of the genus as a whole (for a review, see ref. 2). Rapid advances in DNA sequence analysis have made these chemical agents much more amendable to systematic interspecific analysis than any comparably diverse set of natural products produced by any genus or family of animals or plants.
The analysis carried out on Conus venom peptides suggests that a majority of the estimated >50,000 peptides are encoded by only ≈12 conotoxin gene superfamilies. These superfamilies have undergone rapid amplification and divergence, accompanying the parallel radiation and diversification of Conus species at a macroevolutionary level (Conus is arguably the most species-rich genus of living marine invertebrates). Each major Conus peptide gene superfamily comprises thousands of genes, encoding different peptides. This leads to the remarkable functional diversity seen among the ≈50,000 different peptides. A majority of all Conus peptides exert a powerful effect on some specific ion channel or receptor target (4). It is fair to say that the snails likely have evolved a greater diversity of ion channel-targeted pharmacological agents than even the largest of pharmaceutical companies (this diverse array includes peptides that are being developed for use as human pharmaceuticals). These venom peptides have allowed different cone snail species to specialize on at least five different phyla of prey and defend themselves against a spectrum of predators that might be even more diverse.
Most protein genes initially produce a translation product that is >100 amino acids in length, a size sufficient to allow conventional folding by multiple intramolecular interactions. Toxin proteins found in venoms are generally smaller (50–100 aa), with additional stability provided by disulfide bonds. In effect, the cone snails have extended this tendency one step further, with some venom peptide superfamilies being the smallest highly structured but functionally diverse classes of gene products known (12–20 aa with two to three disulfide bonds). Conopeptide evolution has resulted in a large diversity of biological function being generated in each conotoxin superfamily. Thus, Conus peptide superfamilies are like other major classes of proteins produced through gene translation: structural and functional novelty can evolve, and thus, conotoxins are in many respects “microproteins” and differ from more conventional unstructured peptides.
Each conotoxin superfamily has a characteristic arrangement of cysteine residues, which is assembled into a particular disulfide bonding configuration (the “disulfide framework”). The latter is the primary determinant of polypeptide backbone structure. Despite hypermutation of the amino acids between Cys residues, the disulfide framework generally remains conserved within a superfamily, generating a characteristic scaffold. In principle, for peptides with six Cys residues (characteristic of at least four conotoxin superfamilies) there are 15 different disulfide-bonded arrangements possible, with only one being biologically relevant. How does folding to this single structural framework (instead of the 14 other possibilities) occur? This is a specialized version of the more general protein folding problem, one of the central unsolved questions in biology.
Efficient solutions to this specialized folding problem must have been evolved by cone snails before they could effectively explore (in an evolutionary sense) the alternative world of microproteins that they have so efficiently exploited for their diverse physiological purposes. In this article, data are presented that are relevant to this previously unaddressed issue. There are two levels at which solutions to the specialized folding problem of Conus peptides could have evolved: (i) specialized intramolecular interactions may stabilize conformation, and (ii) intermolecular interactions with extrinsic factors, perhaps within the endoplasmic reticulum (ER), may promote appropriate folding pathways within the ER. We explore both possibilities.
An unusual feature of Conus venom peptides is the high degree of posttranslational modification observed (5). We demonstrate that posttranslational modification can generate intramolecular interactions that facilitate conopeptide folding. For potential intermolecular interactions, we show that multiple isoforms of protein disulfide isomerase (PDI), which catalyze protein thiol disulfide exchange reactions, are present within Conus venom ducts, and we show that, apart from the conotoxins themselves, these isoforms are the major soluble protein components of the ducts.
Materials and Methods
Cloning of Conus PDI. Full-length Conus textile PDI cDNAs were isolated by RT-PCR of venom duct RNA, using degenerate primers based on the amino acid sequence of the highly conserved thioredoxin-like active site motif found in PDI proteins isolated from other organisms. PCR products were gel-purified and cloned into a plasmid vector, and several cloned isolates were sequenced. The DNA sequence of the cloned PCR product contained a long ORF with significant homology to PDI proteins from other organisms. The DNA sequence of this internal PCR product was used to design nested PCR primers for 5′ and 3′ RACE procedures to isolate the full-length cDNA. C. textile venom duct cDNA was synthesized with 5′ and 3′ RACE adapters (Ambion, Austin, TX) and used for RACE amplifications. Specific 5′ and 3′ RACE products were gel-purified, cloned into a plasmid vector, and sequenced. The sequences of each of these RACE products overlapped with the previously isolated central portion of the C. textile PDI cDNA and together these three PCR-generated cDNAs could be merged to give the full-length cDNA sequence encoding the complete PDI protein. To detect PDI expression in venom ducts of other Conus species, RT-PCR was performed in the following species: C. textile, Conus stercusmuscarum, Conus aurisiacus, Conus consors, Conus betulinus, and Conus omaria. The PCR amplification was performed by using primers based on the thioredoxin active site sequence. Approximately 20 ng of venom duct cDNA and Taq polymerase was used for the PCRs.
Protein Analysis of Venom Ducts. The venom duct from C. textile was dissected and immediately divided into four equal parts. Each part of the venom duct was ground under liquid nitrogen. Extraction was performed in 1 ml of 10 mM Tris·HCl, pH 7.8 containing 0.25 M sucrose and 5 mM EDTA at 4°C. After homogenization, the solution was centrifuged. The 50 μl of resulted supernatant was lyophilized and dissolved in 30 μl of SDS-electrophoresis buffer, boiled for 5 min, and applied on 4–20% Tris-glycine gel. Proteins were then electroblotted onto Immobilion poly(vinylidene difluoride) membrane (0.45 μm) (Millipore) for 1 h at 50 V. Proteins were visualized by using Coomassie blue staining, and the protein band of 55 kDa was cut out from the membrane. Amino acid sequencing was performed by the Edman degradation method. After protein separation, polyacrylamide gel was also stained with Coomassie blue.
Peptide Synthesis and Folding. Peptides were synthesized on solid support by using standard Fmoc [N-(9-fluorenyl)methoxycarbonyl] chemistry. All cysteines were protected with S-trityl groups. The peptides were removed from the resin and purified by using a semipreparative reversed-phase C18 HPLC column in a linear gradient of acetonitrile. After purification, the linear peptide was lyophilized. Folding reactions were initiated by injecting a resuspended linear peptide into a 200-μl solution containing 0.1 M Tris·HCl (pH 8.7), 1 mM oxidized glutathione, and 2 mM reduced glutathione. The folding reactions contained CaCl2 or MgCl2 at concentrations indicated in the text or 1 mM EDTA. The final peptide concentration was 20 μM. After an appropriate folding time, the reactions were quenched by acidification with formic acid (10% final concentration). The samples were separated by reversed-phase C18 analytical HPLC in the gradient of acetonitrile (from 9% to 31.5% acetonitrile in 0.1% trifluoroacetic acid). The accumulation of the native form was calculated relative to other folding species from integration of the HPLC peaks.
Assays for Biological Activity of Synthetic Peptides. The activities of synthetic peptides were evaluated by using 13- to 14-day-old mice, weighing 5.9–6.4 g; the intracranial injections were carried out as described (6).
Results
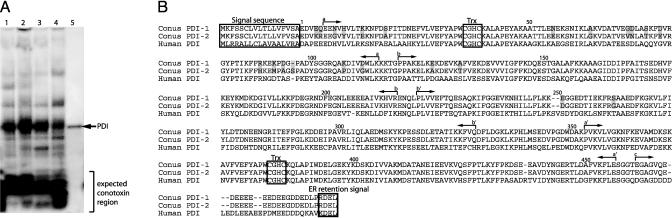
PDI Is the Major Soluble High Molecular Weight Polypeptidic Component of Conus Venom Ducts. The oxidative folding of secreted proteins with disulfide bonds is thought to be promoted by PDIs (7, 8). These enzymes contain four thioredoxin-like domains, two of which have the sequence –CXXC–, that are believed to catalyze formation and isomerization of protein disulfide bonds. The unprecedented density of disulfide linkages in conotoxins (up to 50% of all amino acids can be Cys residues involved in disulfide bonding) led us to initiate the characterization of PDI from cone snail venom ducts. Protein extracts from C. textile venom ducts were prepared (see Materials and Methods), and the crude extracts were analyzed by SDS/PAGE. As shown in Fig. 2A, the expected low molecular mass gene products consistent with conotoxins are prominently stained on the gel, but in addition, a major protein band with a molecular mass of ≈55 kDa stands out. This molecular mass is consistent with that of PDIs characterized from other organisms (see marker band in Fig. 2 A).
Fig. 2.
PDI from C. textile venom duct. (A) SDS/PAGE of extracts from different parts of a venom duct. Lanes 1–4 represent equal fragments from distal to proximal parts of the venom duct. Lane 5 is a bovine PDI. The ≈55-kDa band from lane 3 was extracted, and its N-terminal sequence was determined (see text). (B) Structures of PDI-1 and PDI-2 from C. textile. The signal sequence, thioredoxin active sites (Trx), and the ER retention signal are marked. Domains a, b, b′,a′, and c are based on the assignments for human PDI (9, 10). The shaded residues represent differences between PDI-1 and PDI-2.
The identity of the major polypeptide in the 55-kDa band was investigated by microsequencing: a clear N-terminal sequence, –EEVEEQENVY–, was obtained; as will be discussed below, this sequence is consistent with the conclusion that the ≈55-kDa band is PDI, possibly a mixture of different isoforms. Thus, apart from the conotoxins themselves, PDI appears to be the major soluble protein in Conus venom ducts. Such a high level of PDI along the whole length of the duct is consistent with conotoxin synthesis in both distal and proximal portions of the duct and with a major role for PDI in the oxidative folding of conotoxins.
PDI expression was further evaluated in a variety of Conus species by using a PCR screen, with primers matching conserved sequences in PDIs (see Materials and Methods). The PCR fragment obtained from cDNA prepared from a variety of different Conus species representing the major feeding types of the genus, including C. textile, C. stercusmuscarum, C. aurisiacus, C. consors, C. betulinus, and C. omaria (see Fig. 1) was similar in size and intensity to that obtained from C. textile. To characterize Conus PDIs further, we analyzed a cDNA library from C. textile, the cloth-of-gold cone. The sequences of two full-length cDNA clones encoding two different PDI isoforms, PDI-1 and PDI-2, were determined (Fig. 2B). There is a high degree of sequence identity between the two isoforms, but there are two regions of high divergence: amino acids 4–19 (40% sequence divergence) and amino acids 71–96 (32% divergence). Both proteins are ≈75% homologous to the well characterized human PDI sequence shown in Fig. 2.
The cleavage site for the removal of the signal sequence was identified from the microsequencing results for the mature protein detected in the SDS gel. Based on homology to the human PDI, five domains can be distinguished: a, b, b′, a′, and c (9, 10). Two of these (a, a′) are domains that have sequence similarity to thioredoxin (a small protein cofactor in ribonucleotide reduction), whereas two (b, b′) show little sequence similarity but have a thioredoxin-like fold. The two putative active-site –CGHC– sequences are located in domains a and a′; thus, the two regions of high divergence sandwich the –CGHC– site of the a domain. The C-terminal domain is characterized by a high degree of negatively charged Asp and Glu residues, postulated to serve as the Ca2+ binding domain (11, 12). The C-terminal sequence RDEL– is a standard ER retention signal. Thus, the Conus PDI isoforms cloned contain all characteristic features of PDIs from other organisms. The results with the directly sequenced protein band in Fig. 2 A suggest that there may be additional venom duct PDI isoforms. Preliminary molecular evidence for additional PDI isoforms has been independently obtained (I.G. and J.E.G., unpublished results). We are presently defining all Conus PDI isoforms and have expressed the two shown in Fig. 2B. Enzymatic properties of the expressed Conus PDIs need to be assessed to determine whether these may exhibit specificity for particular conotoxin substrates, relative to other Cys-rich polypeptides.
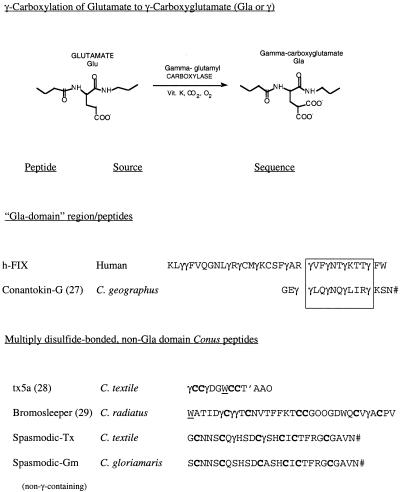
A Role for γ-Carboxyglutamate in Conotoxin Folding. A Conus peptide recently characterized from C. textile, the “spasmodic peptide” (6), has serendipitously presented an opportunity to evaluate the function of an unusual posttranslational modification found in Conus peptides, the conversion of glutamate to γ-carboxyglutamate (see Fig. 3). A closely homologous peptide identified by RT-PCR from a closely related species, the glory-of-the-sea cone, Conus gloriamaris (see Fig. 1) exhibited considerable sequence identity (24/27 amino acids identical) to the C. textile peptide (13). However, no γ-carboxyglutamate residues are present in the C. gloriamaris peptide. Thus, two very closely related natural peptides from different Conus species are largely identical in sequence, but one has two γ-carboxyglutamate residues, whereas the other has none (see Fig. 4 a and b).
Fig. 3.
Comparison of Gla-containing peptides from Conus species. The posttranslational modification of glutamate to γ-carboxyglutamate (Gla or γ) is shown, and a comparison of the sequences of four Gla-containing Conus venom peptides to the N-terminal sequence of human blood clotting factor IX is shown. Note that one Conus peptide, conantokin-G from C. geographus, has a Gla domain motif that has similar spacing to factor IX (see boxed sequences), whereas the three other Gla-containing peptides do not have an arrangement of Gla residues characteristic of a Gla domain. #, amidated C terminus; W, 6-bromotryptophan; T′, glycosylated threonine; γ, γ-carboxyglutmate; h-FIX, human factor IX.
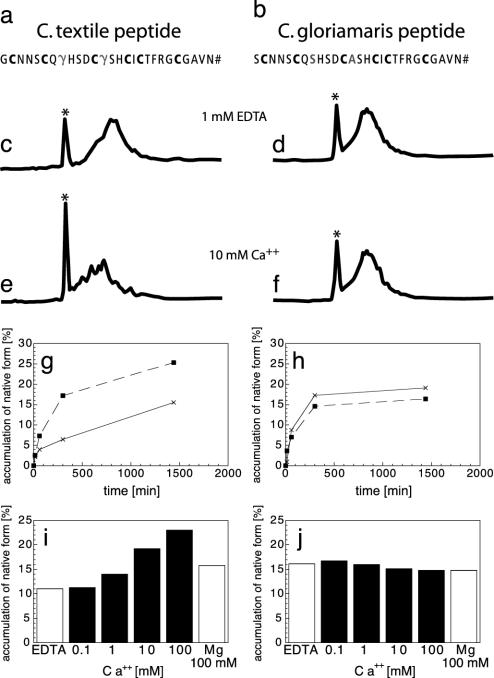
Fig. 4.
Effect of Ca2+ on the oxidative folding of peptides from C. textile and C. gloriamaris. (a and b) Sequences of peptides from C. textile and C. gloriamaris.(c–f) HPLC analysis of oxidative folding of the peptides. The folding mixtures contained reduced and oxidized glutathione, and the reactions were carried out in the presence of 10 mM CaCl2 or 1 mM EDTA. Experimental details are described in Materials and Methods. The correctly folded species (*) has the shortest retention time. (c) Folding of C. textile peptide in the absence of Ca. (d) Folding of C. gloriamaris peptide in the absence of Ca. (e) Folding of C. textile peptide in the presence of Ca. (f) Folding of C. gloriamaris peptide in the presence of Ca. (g and h) Kinetics of forming the native peptides from C. textile (g) and C. gloriamaris (h). The folding reactions were performed as described in Materials and Methods. Dashed lines, +10 mM Ca2+; solid lines +1 mM EDTA. (i and j) Accumulation of the native peptides from C. textile (i) and C. gloriamaris (j) in the folding reactions at 24 h, with increasing concentration of calcium ions. Two other bars represent accumulation of the native peptides in the presence of 1 mM EDTA or 100 mM Mg2+. All experimental points are averages from duplicate experiments.
Both the C. gloriamaris and C. textile peptides were chemically synthesized on solid support and folded in the presence of glutathione as described in Materials and Methods. The biological activities of the two purified synthetic peptides were evaluated by intracerebroventricular injection into mice; despite the differences in γ-carboxylation, no difference in biological activity could be detected. When injected into mice, both peptides elicited the response characteristic of the native spasmodic peptide, hypersensitivity to touch at a dose of 2 nmol and hyperactivity and convulsions at 5 nmol. These results indicate that the Gla residues do not play a major role in the molecular interactions between the mature folded peptide and the physiological target of the C. textile peptide.
We then assessed whether the Gla residues might affect peptide folding. Under our standard Ca-free folding conditions, yields of the properly folded C. gloriamaris peptide were slightly higher than for C. textile peptide (Fig. 4 c and d). Because γ-carboxyglutamate residues chelate Ca2+, we evaluated the effects of Ca2+ on the folding of the two peptides. As shown in Fig. 4e, the addition of Ca2+ results in a significant increase in the yield of properly folded C. textile peptide; strikingly, Ca2+ elicited no increase in the yield of the C. gloriamaris peptide, which lacks γ-carboxyglutamate (Fig. 4f). Above 10 mM Ca2+, the C. textile (Gla+) peptide is more efficiently folded than the C. gloriamaris (Gla-) peptide, a reversal of the situation in the absence of Ca2+ (Fig. 4 g and h). Furthermore, the magnitude of the increase in yield of correctly folded peptide is not observed when Mg2+ is added over the same concentration range (Fig. 4 i and j). Moreover, Ca2+ at 10 mM concentration did not significantly change the reductive unfolding kinetics of C. textile (Gla+) peptide (data not shown). These results are consistent with the conclusion that once the peptides are correctly folded, γ-carboxyglutamate residues make no significant contribution to biological activity or stability; however, the γ-carboxyglutamate residues clearly facilitate oxidative folding when Ca2+ is present, as would be the case in the ER under in vivo folding conditions.
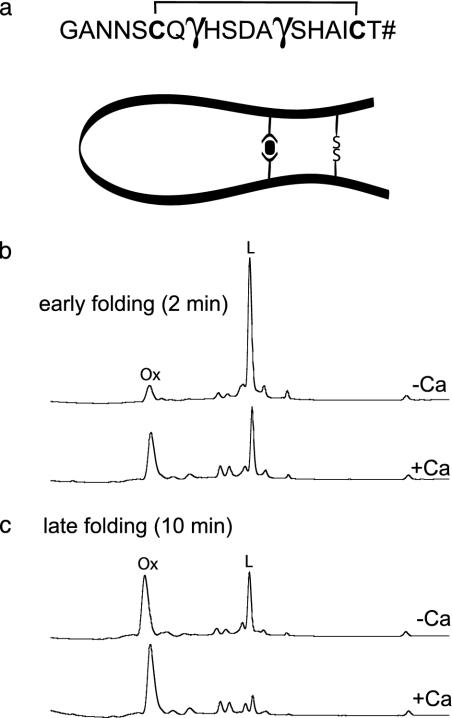
To further investigate a mechanism of the Ca2+-assisted oxidative folding of the Gla-containing peptide, we synthesized a model peptide, which represents a fragment of the conotoxin from C. textile, but containing only one pair of cysteines and two Gla residues (Fig. 5a). The second pair of Cys was replaced by Ala. The rationale for using this model peptide is to assess whether the presence of the Gla residues accelerates formation of a disulfide bond in the presence of Ca2+. As shown in Fig. 5 b and c, the presence of 10 mM Ca2+ in the reaction mixture increased the accumulation of the oxidation product both at early (2 min) and later (10 min) time points. These results provide a mechanistic rationale, at least in part, for the observed effects of Ca2+ on folding of the conotoxin from C. textile (see Discussion).
Fig. 5.
Ca2+-assisted oxidation of the Gla-containing model peptide. The peptide (a) was designed based on the sequence of peptide from C. textile. Cys-2, Cys-12, and Cys-16 were replaced by Ala residues, and Thr-19 had an amidated C terminus (labeled #). The disulfide bond between Cys-6 and Cys-18 is also shown. A hypothetical model illustrating how a coordination of Ca2+ by the two Gla residues in the peptide may favor formation of a disulfide bridge (-SS-) by bringing two Cys thiols in close proximity. (b and c) HPLC analysis of oxidation of the model peptide with a mixture of oxidized (1 mM) and reduced (2 mM) glutathione and in the presence of either 10 mM CaCl2 or 1 mM EDTA. The oxidation mixtures were quenched by acidification after 2 or 10 min and analyzed by reversed-phase C18 analytical HPLC. L, linear (reduced) peptide; Ox, oxidized form.
Discussion
In the course of their evolutionary history, the venomous cone snails have developed an unprecedented number of diverse, highly structured peptides that are expressed in their venom ducts. These peptides as a class are the smallest ribosomally translated functional gene products (microproteins). Most of these peptides potently interfere with the function of a specific ion channel target. To have the potent and selective pharmacological activity required, the peptides have to be properly folded with the correct disulfide connectivity. Clearly, cone snails have generated mechanisms to facilitate small-peptide folding; this article begins to address such mechanisms. There are at least two motivations for embarking on this general research direction: the first (rather pedestrian) reason is that understanding factors that facilitate proper conotoxin folding in vivo should help in the routine production of properly folded Conus peptides in vitro. However, our long-term hope is that by elucidating how cone snails are able to fold these small peptides properly, we will also gain insights into the more general problem of protein folding.
All Conus peptide precursors have highly conserved N-terminal signal sequences that direct the newly translated precursor to the ER membrane, transferring it from a highly reducing, low Ca2+ environment (the cytosol) to an oxidizing, high Ca2+ environment (the ER matrix). In the oxidizing environment of the ER (14), the high density of cysteine residues in most Conus peptides would be expected to lead to rapid formation of disulfide bonds. We provide evidence for high levels of multiple PDI isoforms in Conus venom ducts; these enzymes generally regulate the formation of disulfide bonds and prevent them from being formed randomly. PDI appears to be the most highly expressed high molecular weight polypeptide component of Conus venom ducts. PCR amplification of PDI is only intermittently successful when other Conus tissues are similarly analyzed, presumably because of a much lower frequency of PDI transcripts. These observations are consistent with a key role for PDI in the folding of the major secreted products of these cells, the conotoxins. An obvious possibility to be explored is whether different Conus PDI isoforms have evolved specialized features to facilitate proper folding of the specific disulfide frameworks in the various conotoxin superfamilies. An additional question is whether Conus PDIs have synergistic interactions with other specialized chaperones in the ER that could lead to even greater facilitation of conotoxin folding.
The many posttranslational modifications observed in Conus peptides are believed to enhance the affinity and/or pharmacological specificity of modified conopeptides; however, the results presented above suggest an additional function for some Conus peptide posttranslational modifications: to facilitate proper folding. In the example presented (γ-carboxylation of glutamate residues), formation of the correctly disulfide-bonded conopeptide was enhanced by the Gla residues in the presence of physiological concentrations of Ca2+, but did not appear to be important for activity of the folded peptide. The results presented suggest that the presence of γ-carboxyglutamate residues facilitate folding of the C. textile peptide in vivo.
A role for Ca2+ in oxidative folding was previously suggested for secreted proteins such as the low-density lipoprotein (LDL) receptor (15, 16), Notch1 receptors (17) or α-lactalbumin (18). For the LDL receptor, a “calcium box” rich in Asp or Glu was postulated to direct folding by chelating Ca2+ and nucleating early folding intermediates as the LDL receptor entered the ER. Similarly, as disulfide-rich γ-carboxylated Conus peptides are translocated through the ER membrane, we suggest that a compact calcium mini-box with γ-carboxyglutamate residues may either facilitate a favorable spatial orientation of Cys residues for correct disulfide bond formation at early stages of folding, leading to the final biologically active conformation, or alternatively, inhibit formation of undesirable disulfide bonds. The experiment on a model peptide (Fig. 5) provides support for the former, but does not eliminate the possibility that the latter may contribute as well. γ-Carboxylation does not appear to be required for the biological activity of the mature, properly folded peptide gene product, nor is it required for maintaining the structure of peptide once correct disulfide bonds are formed.
γ-Glutamyl carboxylases, the enzymes that convert glutamate to γ-carboxyglutamate (Gla), were previously shown to be highly conserved, clearly predating the divergence of vertebrates, arthropods, and molluscs (19). Thus, an ancestral role (more general than the specialized uses of the modification in blood clotting in mammals and venom peptides in Conus) that provided selective pressure for enzyme conservation as animal phyla diverged is strongly indicated. The present results raise the possibility that the proper folding of proteins, directed by the presence of Gla residues at strategic loci in the polypeptide, may have been that ancestral role. We suggest that this putative ancient role of Gla was recapitulated to facilitate folding of some of the very rapidly evolving, unusually small Conus peptides.¶ Such a folding mechanism for proteins may have been more generally important earlier in evolution, but it was probably largely supplanted later by other mechanisms for facilitating folding of larger polypeptides, such as specialized molecular chaperones (20, 21).
The role of Gla suggested above provides an attractive general mechanism for folding small polypeptides, perhaps even including the primordial proteins. Based on structural work on signal recognition particle peptides, it was recently suggested (22) that the first proteins evolved as membranes formed, when RNA still dominated biochemistry. Specifically, the first functional polypeptide-like chains in incipient life forms were created to deal with a membrane surrounding the catalytic/informational RNA. If this view is correct, then the possibility is raised that γ-carboxyglutamate, with its capacity both for interacting with membranes and directing folding may have been present in the earliest functional polypeptides, which were presumably much smaller than present-day proteins. Once a Ca2+-free cytosol evolved, however, a doubly negatively charged amino acid might become a liability for intracellular protein function, and in most taxa at the present time, γ-carboxyglutamate is probably largely a relict amino acid in a few secreted proteins. This modification remains prominent only in those present-day phylogenetic systems where more specialized uses have evolved (such as mammalian blood clotting and Conus venom peptides) (Fig. 3).
The canonical organization of conopeptide precursors, into an N-terminal signal sequence, an intervening pro region, and a single copy of the mature toxin region at the C terminus was first established a decade ago (23). The striking contrast found between the conserved signal sequence and propeptide regions versus the hypermutable mature toxin region led to the suggestion that the conserved regions of the precursor were essential for proper folding “such that the single biologically active disulfide-bonded configuration can be specifically formed.” It was pointed out in the same paper that in vivo, additional factors to facilitate the folding process might be required, such as PDI; in this work, we provided the characterization of PDI from Conus venom ducts. Evidence that posttranslational modification enzymes may also be extrinsic factors that facilitate proper folding was presented above. For both PDI and posttranslational modification enzymes, the propeptide region may play a key role by providing anchor sites, so that these factors can act efficiently on the mature toxin region. This has been directly demonstrated for the posttranslational modification enzyme that carboxylates glutamate to γ-carboxyglutamate, a specific sequence present in the conotoxin precursor propeptide region is recognized by the enzyme (24). Evidence that PDIs facilitate proper folding of a conopeptide precursor was also recently obtained (O.B. and G.B., unpublished results). Thus, the relatively conserved propeptide regions may have a function in establishing the specific disulfide bonding conformation of Conus peptides.
The high expression levels and multiple isoforms of PDI in Conus venom ducts and the involvement of a posttranslational modification enzyme are by no means the only adaptation that cone snails have made to efficiently fold the small, highly structured, rapidly evolving peptides in their venoms. The studies described here only serve to identify candidates in what is likely a long list of molecular adaptations necessary for the genus Conus to implement their unusual biochemical/pharmacological strategy for interacting with the prey, predators, and competitors in their environment.
In closing, we address the link between the amplification and diversification of conotoxins and macroevolutionary trends in Conus. The fossil record indicates that the genus had its origins after the Cretaceous extinction, with a modest initial radiation in the Eocene (25); molecular data are consistent with several early lineages of Conus diverging from each other at this time (2, 26). Together, the fossil record and molecular data suggest that one of these early Conus lineages expanded rapidly at the beginning of the Miocene, a radiation that basically continues to the present day. This gave rise to the great majority of the shallow-water species, including all of the fish-hunting and mollusc-hunting Conus.
The peptidic nature of conotoxins will make it possible to assess general evolutionary patterns in the conotoxin genes as these radiations of Conus lineages took place. However, we can begin to evaluate a different level of molecular adaptations as well. As we understand more about the oxidative folding process, we should in principle be able to determine at which point the various components that facilitate oxidative folding of conotoxins appear. Thus, one question that can be evaluated through a comparison of different early diverging lineages is whether most of these adaptations took place early in the history of the genus, or whether a breakthrough in folding smaller conotoxins could have been a factor that made the major radiation of cone snails in the Miocene possible.
Acknowledgments
This work was supported by National Institute of General Medical Sciences Grant GM 48677.
This paper results from the Arthur M. Sackler Colloquium of the National Academy of Sciences, “Chemical Communication in a Post-Genomic World,” held January 17–19, 2003, at the Arnold and Mabel Beckman Center of the National Academies of Science and Engineering in Irvine, CA.
Abbreviations: PDI, protein disulfide isomerase; ER, endoplasmic reticulum.
Footnotes
An obvious question is why γ-carboxyglutamate in C. textile peptide is not present in C. gloriamaris peptide. C. textile peptide is from a tropical Conus species from a warm, shallow-water habitat. C. gloriamaris peptide is from a deep-water species (≈100 fathoms); the significantly cooler habitat may minimize the pressure to facilitate folding by the presence of Gla. An alternative/additional rationale is that C. textile peptide is a major venom component, whereas C. gloriamaris peptide was never directly detected in venom, but only from a cDNA clone (implying that it may be a quantitatively minor venom component). A higher production rate would provide additional selective pressure for mechanisms that facilitate folding.
References
- 1.Olivera, B. M., Rivier, J., Clark, C., Ramilo, C. A., Corpuz, G. P., Abogadie, F. C., Mena, E. E., Woodward, S. R., Hillyard, D. R. & Cruz, L. J. (1990) Science 249, 257-263. [DOI] [PubMed] [Google Scholar]
- 2.Olivera, B. M. (2002) Annu. Rev. Ecol. Syst. 33, 25-42. [Google Scholar]
- 3.Röckel, D., Korn, W. & Kohn, A. J. (1995) Manual of the Living Conidae (Verlag Christa Hemmen, Wiesbaden, Germany).
- 4.Terlau, H. & Olivera, B. M. (2004) Physiol. Rev. 84, 40-67. [DOI] [PubMed] [Google Scholar]
- 5.Craig, A. G., Bandyopadhyay, P. & Olivera, B. M. (1999) Eur. J. Biochem. 264, 271-275. [DOI] [PubMed] [Google Scholar]
- 6.Lirazan, M. B., Hooper, D., Corpuz, G. P., Ramilo, C. A., Bandyopadhyay, P., Cruz, L. J. & Olivera, B. M. (2000) Biochemistry 39, 1583-1588. [DOI] [PubMed] [Google Scholar]
- 7.Goldenberger, R. F., Epstein, C. J. & Anfinsen, C. B. (1963) J. Biol. Chem. 238, 628-635. [PubMed] [Google Scholar]
- 8.Tu, B. P., Ho-Schleyer, S. C., Travers, K. J. & Weissman, J. S. (2000) Science 290, 1571-1574. [DOI] [PubMed] [Google Scholar]
- 9.Kemmink, J., Darby, N. J., Dijkstra, K., Nilges, M. & Creighton, T. E. (1997) Curr. Biol. 7, 239-245. [DOI] [PubMed] [Google Scholar]
- 10.Darby, N. J., Penka, E. & Vincentelli, R. (1998) J. Mol. Biol. 276, 239-247. [DOI] [PubMed] [Google Scholar]
- 11.Macer, D. R. & Kock, G. L. (1988) J. Cell Sci. 91, 61-70. [DOI] [PubMed] [Google Scholar]
- 12.Lucero, H. A. & Kaminer, B. (1999) J. Biol. Chem. 274, 3243-3251. [DOI] [PubMed] [Google Scholar]
- 13.Miles, L. A., Dy, C. Y., Nielsen, J., Barnham, K. J., Hinds, M. G., Olivera, B. M., Bulaj, G. & Norton, R. S. (2002) J. Biol. Chem. 277, 43033-43040. [DOI] [PubMed] [Google Scholar]
- 14.Gilbert, H. F. (1990) Adv. Enzymol. Relat. Areas Mol. Biol. 63, 69-172. [DOI] [PubMed] [Google Scholar]
- 15.Bieri, S., Atkins, A. R., Lee, H. T., Winzor, D. J., Smith, R. & Kroon, P. A. (1998) Biochemistry 37, 10994-11002. [DOI] [PubMed] [Google Scholar]
- 16.Koduri, V. & Blacklow, S. C. (2001) Biochemistry 40, 12801-12807. [DOI] [PubMed] [Google Scholar]
- 17.Aster, J. C., Simms, W. B., Zavala-Ruiz, Z., Patriub, V., North, C. L. & Blacklow, S. C. (1999) Biochemistry 38, 4736-4742. [DOI] [PubMed] [Google Scholar]
- 18.Rao, K. R. & Brew, K. (1989) Biochem. Biophys. Res. Commun. 163, 1390-1396. [DOI] [PubMed] [Google Scholar]
- 19.Bandyopadhyay, P. K., Garrett, J. E., Shetty, R. P., Keate, T., Walker, C. S. & Olivera, B. M. (2002) Proc. Natl. Acad. Sci. USA 99, 1264-1269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gupta, R. S., Aitken, K., Falah, M. & Singh, B. (1994) Proc. Natl. Acad. Sci. USA 91, 2895-2899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gupta, R. S. (1995) Mol. Biol. Evol. 12, 1063-1073. [DOI] [PubMed] [Google Scholar]
- 22.Walter, P., Keenan, R. & Schmitz, U. (2000) Science 287, 1212-1213. [DOI] [PubMed] [Google Scholar]
- 23.Woodward, S. R., Cruz, L. J., Olivera, B. M. & Hillyard, D. R. (1990) EMBO J. 1, 1015-1020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bandyopadhyay, P. K., Colledge, C. J., Walker, C. S., Zhou, L.-M., Hillyard, D. R. & Olivera, B. M. (1998) J. Biol. Chem. 273, 5447-5450. [DOI] [PubMed] [Google Scholar]
- 25.Kohn, A. J. (1990) Malacologia 32, 55-67. [Google Scholar]
- 26.Espiritu, D. J. D., Watkins, M., Dia-Monje, V., Cartier, G. E., Cruz, L. J. & Olivera, B. M. (2001) Toxicon 39, 1899-1916. [DOI] [PubMed] [Google Scholar]
- 27.McIntosh, J. M., Olivera, B. M., Cruz, L. J. & Gray, W. R. (1984) J. Biol. Chem. 259, 14343-14346. [PubMed] [Google Scholar]
- 28.Walker, C., Steel, D., Jacobsen, R. B., Lirazan, M. B., Cruz, L. J., Hooper, D., Shetty, R., DelaCruz, R. C., Nielsen, J. S., Zhou, L., et al. (1999) J. Biol. Chem. 274, 30664-30671. [DOI] [PubMed] [Google Scholar]
- 29.Craig, A. G., Jimenez, E. C., Dykert, J., Nielsen, D. B., Gulyas, J., Abogadie, F. C., Porter, J., Rivier, J. E., Cruz, L. J., Olivera, B. M. & McIntosh, J. M. (1997) J. Biol. Chem. 272, 4689-4698. [DOI] [PubMed] [Google Scholar]