Abstract
Sets of RNA ladders can be synthesized by transcription of a bacteriophage-encoded RNA polymerase using 3′-deoxynucleotides as chain terminators. These ladders can be used for sequencing of DNA. Using a nicked form of phage SP6 RNA polymerase in this study substantially enhanced yields of transcriptional sequencing ladders. Matrix-assisted laser desorption/ionization time-of-flight mass spectrometry (MALDI-TOF MS) of chain-terminated RNA ladders allowed DNA sequence determination of up to 56 nt. It is also demonstrated that A→G and C→T variations in heterozygous and homozygous samples can be unambiguously identified by the mass spectrometric analysis. As a step towards single-tube sequencing reactions, α-thiotriphosphate nucleotide analogs were used to overcome problems caused by chain terminator-independent, premature termination and by the small mass difference between natural pyrimidine nucleotides.
INTRODUCTION
The sequencing of the human genome is scheduled for completion by the end of 2003 (1). This will increase the need for robust and high-throughput technologies for high-fidelity DNA sequencing and genotyping because acquisition of the comprehensive human genome sequence will set the stage for new applications, such as allelic association studies, which exploit genome-wide genetic variations obtained from large populations. This increasing need has led to the development of several new approaches. In some of these, mass spectrometry (MS) has been used instead of the conventional gel electrophoresis for DNA sequencing and variation detection (2,3)
Two mild MS ionization techniques are used for biopolymer analysis: matrix-assisted laser desorption/ionization (MALDI) and electrospray ionization (ESI). Because the MALDI process generates mainly singly charged molecular ions, which are separated by their mass:charge ratios, MALDI time-of-flight (TOF) MS is suitable for the analysis of complex mixtures. The measurement of nucleic acids by MS is extremely quick and requires neither labeling nor staining. Amenability to automation also makes MS suitable for high-throughput analyses. Moreover, MS does not suffer from a common problem in gel-based analysis of compression artifacts due to secondary structures, which lead to misreading of the sequence.
However, MALDI-TOF MS has the weaknesses of (i) compromised resolution, which is highly sensitive to salt formation primarily with alkali ions, and (ii) the low upper limit of mass range. The resolution and sensitivity are severely impaired as mass increases. One possible explanation for the limitation in mass range is the depurination and fragmentation of DNA in the MALDI process. The stabilizing effect of substituting the 2′-hydrogen with electronegative groups, such as hydroxyl and fluorine, has been reported previously (4). A nested set of RNA fragments has been expected to be more suitable for MALDI-TOF MS analysis than DNA counterparts, because RNA shows higher sensitivity and better stability in MALDI MS than DNA (5–7).
Sequencing ladders of RNA can be obtained either by transcriptional synthesis of chain-terminated RNA fragments with occasional incorporation of 3′-deoxynucleotide (3′-dNTP), or by incomplete enzymatic or chemical digestion of RNA to produce a set of shorter RNA fragments. MS analyses of nuclease-digested and chemically-degraded RNA fragments for sequencing of RNA have been reported (6,8,9). However, MS analysis of the chain-terminated RNA fragments for sequencing has not yet been reported. Sets of aborted transcripts that revealed DNA sequence when electrophoretically separated have been obtained by using phage RNA polymerase transcription in the presence of either 3′-dNTPs (10), missing rNTPs (11) or fluorescent dye-labeled 3′-dNTPs (12).
In this study, we synthesized a nested set of chain-terminated RNA fragments with a proteolytically modified SP6 RNA polymerase, which produces a significantly higher yield than the intact polymerase. The RNA ladders allowed sequence determination not only by gel electrophoretic separation but also by mass determination using MALDI-TOF MS. Furthermore, we demonstrate that this analysis is accurate enough to unambiguously identify genotypes of heterozygous alleles of the most common DNA sequence variations, single nucleotide polymorphisms (SNPs).
MATERIALS AND METHODS
Transcriptional sequencing reactions
Bacteriophage SP6 and T7 RNA polymerases and all the nucleotides were purchased from Amersham Pharmacia Biotech, except for the 4-thio-UTP (S4UTP) and the 3′-dNTPs, which were from United States Biochemical and Boehringer Mannheim, respectively. The nicked SP6 RNA polymerase was purified from Escherichia coli strain JM109/pACS6R (13) by the method described previously (14). The SP6 RNA polymerase was cleaved into two fragments during purification from the JM109 cell extract (15,16), just as has been reported for T7 RNA polymerase (17).
Transcription was carried out in 10-µl reactions consisting of 40 mM Tris–HCl, pH 7.9, 6 mM MgCl2, 10 mM dithiothreitol, 2 mM spermidine–HCl, 0.5 mM rNTPs, 2 µCi [α-32P]CTP or [α-32P]UTP (3000 Ci/mmol), 4 U of RNasin (Promega), 0.5–1.0 pmol template and 10 U phage RNA polymerase at 37°C for 30 min. For UMP analog incorporation, 0.5 mM uridine 5′-O-(1-thiotriphosphate), or α-thio-UTP (UTPαS) purchased from Amersham Pharmacia Biotech was added in place of UTP. For transcription reactions with 0.5 mM S4UTP in place of UTP, 40 mM HEPES–KOH (pH 7.2) was used instead of Tris–HCl (pH 7.9) in the transcription buffer.
Transcriptional sequencing reactions were carried out with chain-terminating 3′-deoxynucleotides in the range of 50–100 nM in addition to 0.3 mM rNTPs and 10 µCi [α-32P]CTP or [α-32P]UTP (3000 Ci/mmol). Reactions were stopped by addition of 10 µl EDTA/formamide solution containing 0.025% xylene cyanol FF and 0.025% bromophenol blue, and heated at 90°C for 2 min. The products were analyzed by 12% polyacrylamide 8 M urea gel electrophoresis and quantified by phosphoimaging analysis using a Storm 860 scanner (Molecular Dynamics).
MALDI-TOF MS
Transcription reactions were scaled up to 100 µl without radioactive labeling. RNA products were precipitated by adjusting to 2 M NH4-acetate and addition of 2.5 vol ethanol, and collected by centrifugation. The pellets were washed with 70% ethanol and dried. Desalting was achieved with micro-purification columns as described previously (3). The lyophilized samples of RNA were re-dissolved in 1 µl of pure water, and 0.15 µl was used for MALDI-TOF MS analysis. The matrix solution was a mixture of 3-hydroxypicolinic acid (Aldrich) and 10% (molar ratio) dibasic ammonium citrate, dissolved in 20% acetonitrile. The final concentration of 3-hydroxypicolinic acid was 0.28 M. Approximately 0.15 µl of the matrix solution was loaded onto a stainless steel target and left to dry. Then, 0.15 µl of the analyte solution was added to the matrix spot, and re-crystallized matrix–analyte complex was used directly in linear TOF MS with Voyager DE (PerSeptive Biosystem).
In all cases, target voltage and extraction grid were kept at 18.2 kV when the nitrogen laser was fired. After a delay of 300 ns, the target voltage was raised to 20 kV. The ion guide in the flight tube was kept at –2 V. With such settings, the instrument usually provides mass resolution of ∼700–1000 in the 6–10 kDa mass range for oligonucleotides.
RESULTS
DNA sequencing by chain-terminated RNA fragments
In pursuit of sequencing-by-synthesis of a nested set of RNA fragments, transcription reactions were performed on a promoter-containing plasmid with phage SP6 or T7 RNA polymerase and 3′-dNTPs to abort transcription elongation at specific residues. The radioactively labeled RNA fragments were resolved by sequencing gel electrophoresis to yield sequencing ladders of RNA. A proteolytically nicked form of the SP6 RNA polymerase produced ∼10-fold more RNA fragments than the intact polymerase (Fig. 1). This particular plasmid template contained a terminator sequence of hairpin-independent type, which is recognized by the intact polymerase but not by the nicked form (15). The sum of the run-off and terminated transcripts synthesized by the intact polymerase in a standard multi-round transcription reaction (Fig. 1, lane 1) was similar to the amount of run-off transcripts synthesized by the nicked RNA polymerase under the same conditions (Fig. 1, lane 10). This indicates that the same transcriptional activity of RNA polymerase has been used in the transcription reactions in lanes 1–10 (Fig. 1). When a chain-terminating 3′-dNMP is incorporated at the 3′-end of growing transcript, the complex is arrested and the intact polymerase is not released (15,18). The increased yields of ladders by the nicked polymerase, compared with those by intact polymerase (Fig. 1, lanes 6–9 and lanes 2–5, respectively), suggest that it is released from such arrested complexes and recycled for multi-round initiations.
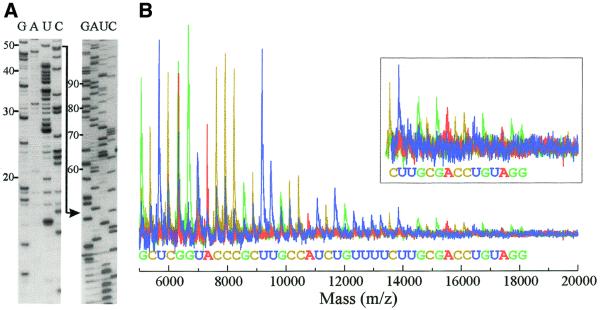
Figure 1.
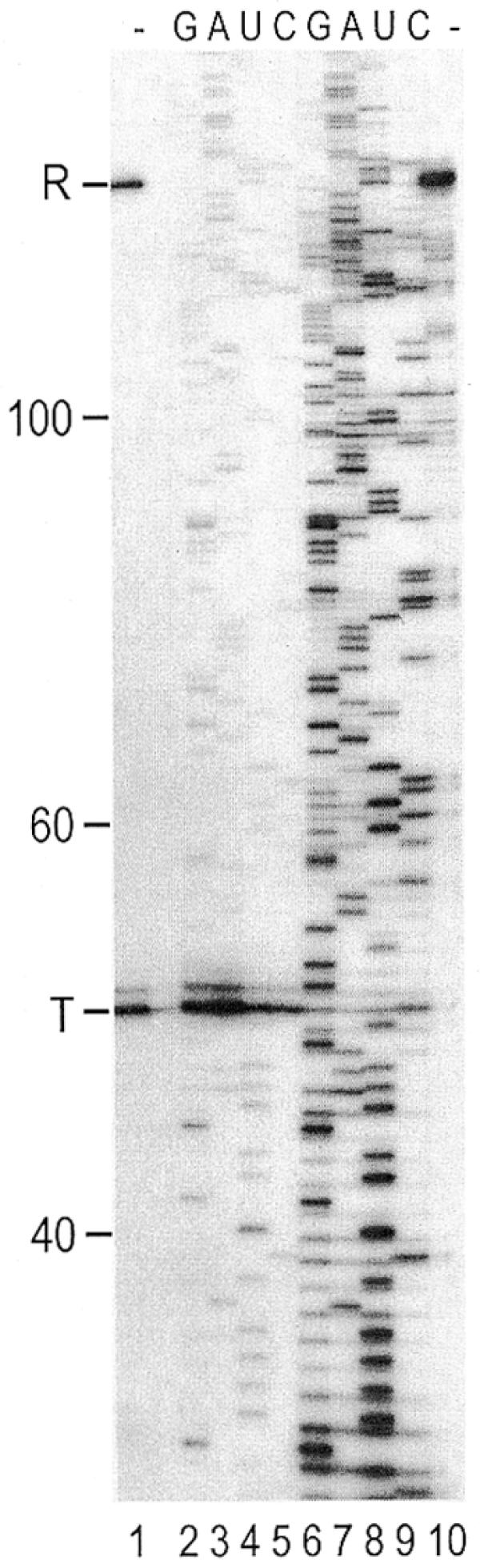
Transcriptional sequencing of DNA by gel electrophoresis. Transcriptional sequencing reactions of the previously described template 1w1 (15) containing a hairpin-independent terminator were carried out with intact (lanes 1–5) and nicked (lanes 6–10) SP6 RNA polymerases. Standard transcription (lanes 1 and 10) and sequencing reaction (lanes 2–9, G, A, U and C) products were labeled with [α-32P]CTP and resolved in parallel using 12% polyacrylamide 8 M urea gel electrophoresis. The nicked RNA polymerase ignores the terminator signal (T), as previously reported (15), and produces only run-off product (R). The size of RNA is shown (in nucleotides) on the left.
The RNA fragments seen were mainly produced by incorporation of 3′-dNMPs and template sequence could be determined up to 300 bp. Some minor bands were also observed across the four sequencing lanes at the same mobility and independent of terminating nucleotide incorporation. These false stops, however, did not severely hamper sequence determination. (See below for nature of the non-specific bands.)
Addition of inorganic pyrophosphatase to sequencing reactions can prevent the pyrophosphorolysis by hydrolyzing pyrophosphates (12,19). The addition of pyrophosphatase to the transcriptional sequencing reactions improved the sequencing band patterns in two aspects. Some non-specific minor bands were suppressed significantly, and some base-specific bands were intensified especially in homo-oligomeric sequence regions (data not shown).
Sequencing and genotyping by MALDI-TOF MS
Four separate reactions, each containing a different 3′-dNTP, were performed with the nicked SP6 RNA polymerase and a plasmid template containing an SP6 promoter with and without a radioactive nucleotide. The radioactively labeled RNA fragments were separated by gel electrophoresis (Fig. 2A). The non-labeled RNA fragments were desalted by a micro-purification column and subjected to MALDI-TOF MS. As estimated from parallel radioactive reactions, 20–1 fmol of 6–18 kDa RNA fragments were used for MS. The mass resolution was 1200–300 in this mass range. Superimposition of four base-specific spectra allowed sequence determination up to the +56 position (transcription start site is +1) by reading major peaks (Fig. 2B).
Figure 2.
Transcriptional sequencing of DNA by MS. Sequencing reactions were carried out with the previously described template 5q10 (15) using nicked SP6 RNA polymerase. The RNA fragments were (A) separated by gel electrophoresis as in Figure 1, and (B) analyzed by MALDI-TOF MS. Base-line corrected spectra are superimposed.
No minor peaks were observed at slightly lighter masses than the major peaks. Thus, 5′-triphosphates appeared to remain intact throughout the process. On the other hand, a few minor peaks appeared at slightly heavier masses than the major peaks. One cause was salt adduction, indicating desalting was not complete. The other cause was elongation abortion products that were generated without terminating nucleotide incorporation. Mass peaks of both 3′-dNTP-dependent and -independent stops were well correlated with RNA bands in denaturing gel electrophoresis. The 3′-end nucleotides of 3′-dNTP-independent aborted products were mostly the correct ones rather than misincorporated ones, based on mass measurements. Thus, the false stops can be distinguished from sequencing peaks by measuring the mass difference between neighboring peaks, and these differences can even reveal the base identity in the high mass resolution range of the spectrum.
A mutation associated with MELAS syndrome was chosen as a model for genotyping. It carries an A→G transition the 3243 position of the human mitochondrial genome. The normal and mutant genes were PCR amplified using an SP6 promoter-containing primer. Also, to mimic a heteroplasmy or heterozygote case, the PCR products of normal and mutant genes were mixed at a 1:1 ratio. The 200-bp PCR products were subjected to G- and C-specific sequencing reactions (Fig. 3).
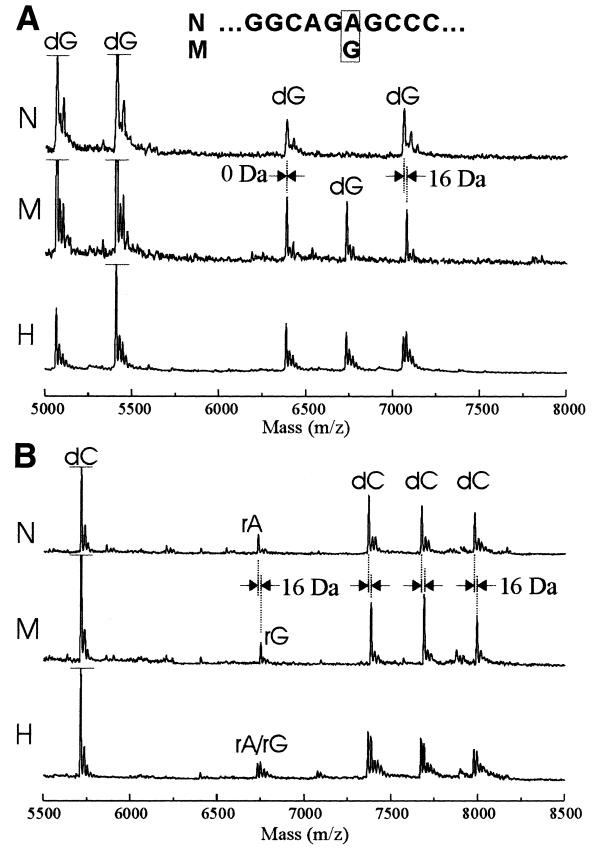
Figure 3.
Mass spectra for the detection of A→G variation. Sequencing reactions of a region involving the MELAS syndrome mutation were performed using nicked SP6 RNA polymerase and analyzed by MALDI-TOF MS. Only a portion of mass spectra of (A) G-specific and (B) C-specific reactions around the mutation is shown. The top (N) and middle (M) spectra in both panels show normal (A-allele) and MELAS (G-allele) samples, respectively. The bottom (H) spectra show a 1:1 mixture of the two samples. The major dG and dC peaks of N, M and H samples in the range 5000–5750 Da are shown as cut off in height (to reduce wasted background space in the graphs), except for the dG peak of H sample near 5000 Da position.
A G- or C-specific peak was not shown at the mutation site from the normal sample (Fig. 3A). On the other hand, a G peak was shown in the MELAS patient sample to reveal the A→G transition and the peaks downstream of it showed a mass increment of 16 Da reflecting the A→G mass difference. The base at the substitution site could also be identified by a 16-Da shift of the C-specific peaks downstream of it (Fig. 3B). In the mixed sample, the G and C peaks downstream of the transition site were doublets with a mass difference of 16 Da.
In addition to the major peaks of 3′-dNMP-incorporated RNA, minor peaks were also observed especially in the C-specific reactions. Mass determination of the minor peaks revealed that their 3′-end carried the rNMP faithfully reflecting the template sequence. Even the minor peaks could be used to confirm the template sequence variations. Thus, these results demonstrate that a sequencing reaction with only one type of 3′-dNTP could be sufficient for the detection of genetic variations in the high-resolution range of MALDI-TOF MS.
DNA sequencing with thio-analog nucleotides
The accuracy and effectiveness of MS could be maximally exploited when the four sequencing reactions are carried out in a single tube and analyzed together. Because the mass difference between U and C is only 1 Da, however, the two reactions used to be separately carried out and analyzed. Using a modified analog of CTP or UTP could result in an increase in this mass difference. We have previously reported that transcriptional sequencing reactions can be carried out with 5-bromo-UTP and 5-iodo-CTP (15). However, the substitution of C5 hydrogen with a more electron-withdrawing group, such as iodine, on the cytosine enhances the liability of the N-glycosidic bond and results in increased fragmentation (4). Peaks would also be broadened due to the natural presence of 79Br and 81Br isotopes.
Here, sequencing reactions were carried out using S4UTP and the Sp isomer of UTPαS in place of UTP (Fig. 4). The mass of the thio-derivatives is 322.2 Da and lies between AMP (329.2 Da) and CMP (305.2 Da). Although sequencing reactions with S4UTP produced RNA ladders under low pH conditions, 3′-dNTP-independent abortion was noticeably increased especially in U-rich regions. Also the elongation complexes are prone to be arrested if they encounter a pause signal, as previously reported (15). These effects appear to be due to the weak stability of S4U:dA base pairing. In contrast, UTPαS was as good a substrate as UTP in sequencing reactions. No increase in minor peaks was observed.
Figure 4.
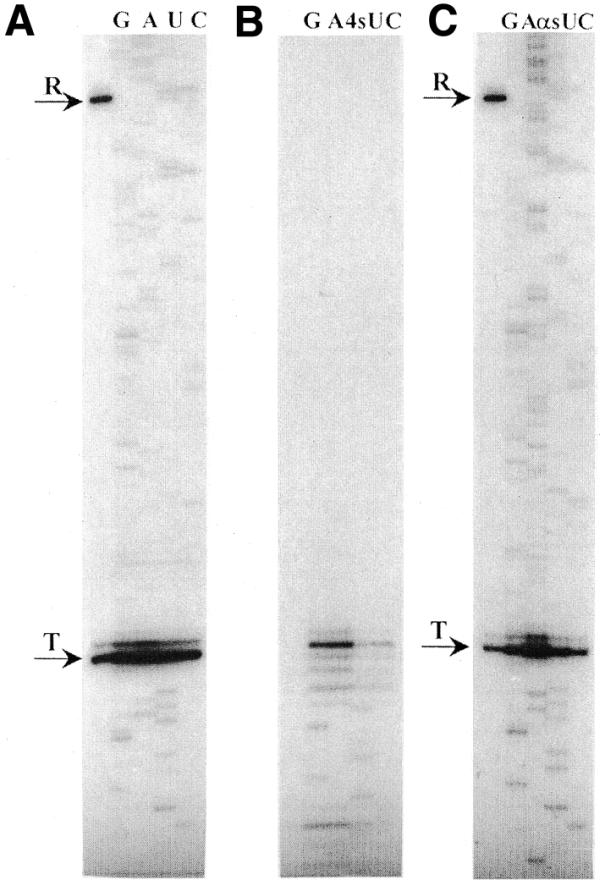
Transcriptional sequencing with thio-analogs of UTP. Sequencing reactions of the template 1w1 (15) with intact SP6 RNA polymerase were carried out using (A) UTP, (B) S4UTP (4sU) or (C) UTPαS (αsU) and the other normal rNTPs. Multi-round transcription reactions of HindIII-digested 1w1 were run in parallel to the left of each panel. See Figure 1 legend.
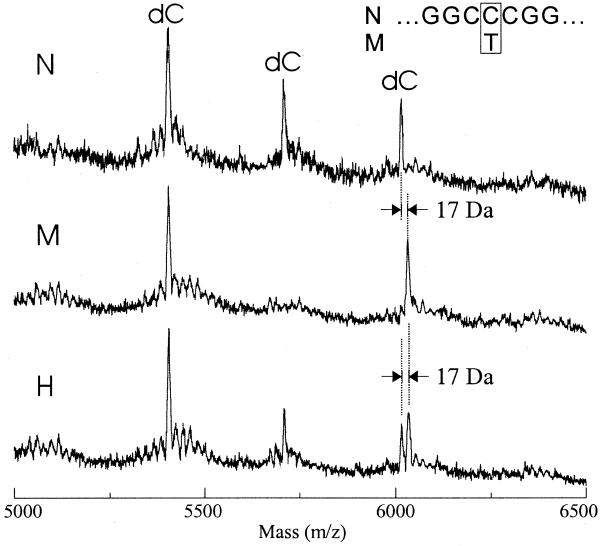
Transcriptional sequencing reactions using UTPαS in place of UTP were subjected to MALDI-TOF MS analysis. Mass peaks of a C-specific reaction with templates containing either C (wild-type) or T (mutant) in the +17 position are shown in Figure 5. The second C peak (∼5.7 kDa position) shown in the wild-type case is absent in the mutant case, revealing the C→T transition. The next C peak (∼6 kDa position) shifted in the mutant case, compared with that in the wild-type case, by 17 Da due to UMPαS incorporation rather than by 1 Da. The increased mass difference thus helped to distinguish between C and U at the variation site. Furthermore, a 1:1 mixture of the two templates was unambiguously distinguished from homogeneous templates by characteristic doublet peaks.
Figure 5.
Mass spectra for the detection of C→T variation. Transcriptional sequencing reactions were carried out with intact T7 RNA polymerase using UTPαS instead of UTP. The templates were derivatives of the previously described plasmid pKJ02 (21) containing a T7 promoter. The TTT sequence at the +16 to +18 positions of pKJ02 was changed to CCC and CTC, and the two resulting templates are different only at the +17 position. Only C-specific reactions analyzed by MALDI-TOF MS are shown. The top (N) and middle (M) spectra show the +17C- and +17T-carrying samples, respectively, and the bottom (H) shows a 1:1 mixture of the two samples.
DISCUSSION
It is demonstrated here that DNA sequence can be determined by MALDI-TOF MS analysis of transcriptional sequencing reactions. The read-length of MS-based RNA sequencing is too short to compete with conventional gel-based DNA sequencing. The throughput of MS-based RNA sequencing is not high enough to be competitive with assays that focus in on individual SNPs. However, the accuracy of MS-based sequencing makes this procedure suitable for identification of genetic variations, not only with DNA ladders (2,3) but also with RNA ladders. An A→G variation was detected unambiguously even in a mixture of the two alleles. RNAs are known to be more stable and sensitive than DNAs in MALDI-TOF MS (4–7). Aberrant primer extension products due to non-specific annealing of a sequencing primer to the template sometimes make base calling ambiguous in DNA replicational sequencing, but are not expected in transcriptional sequencing reactions because only the templates ligated to a promoter are transcribed. Thus, the major application of RNA-based MS sequencing will be for research and diagnostic applications in regions where a complex pattern of SNPs and other variations occur, and where errors cannot be tolerated. Examples include the major histocompatibility complex and various oncogenes.
A serious limitation in RNA-based MS sequencing is due to the small mass difference (1 Da) between U and C, which can be resolved only in the range of low masses. This limitation does not allow MS sequencing of RNA by an exonuclease, nor does it allow for single-tube transcriptional sequencing with the four terminating nucleotides in one tube. The difference of 1 Da was enlarged to 17 Da by using UTPαS in place of UTP, and even a C→T variation in a heterozygote was unambiguously identified by mass determination of sequencing peaks.
One possible drawback with transcription reactions is that RNA polymerase does not possess 3′-exonuclease editing activity so that misincorporation is much more frequent than in DNA replication. This does not seem to be a problem in practice. Even minor products analyzed by MS did not have misincorporated nucleotides, but had ribonucleotides (called ribo peaks here) rather than 3′-deoxynucleotides (called 3′-deoxy peaks here) at the 3′-ends. This premature termination is also observed with primer extension by DNA polymerases. The presence of ribo peaks next to 3′-deoxy peaks potentially reduces resolution. Addition of inorganic pyrophosphatase into the transcription reactions reduced ribo peaks to a certain extent, but the effect was not universal.
Instead of reducing the 3′-rNMP products, one can decrease their mass by 16 Da or increase the mass of 3′-dNMP products by 16 Da, so that the ribo and 3′-deoxy peaks are merged together. When 3′-deoxynucleoside 5′-O-(1-thiotriphosphate) are used instead of 3′-dNTP as chain terminators, all the 3′-deoxy peaks would move downstream by 16 Da to match with the ribo peaks. This approach can also be used in DNA-based MS sequencing, and 2′,3′-dideoxynucleoside 5′-O-(1-thiotriphosphate) can be used as chain terminators. Thus, all the minor MS peaks of premature termination can also be used for base calling.
In this work, the proteolytically modified SP6 RNA polymerase produced a much higher yield of chain-terminated RNA fragments than the intact polymerase. Transcription elongation complexes are generally stable and resistant to dissociation, unless they encounter a termination signal (20), and maintain stability when halted by incorporation of a chain terminator. Because the polymerases are not released from such halted ternary complexes, each polymerase molecule can produce just one RNA molecule at best. On the other hand, the nicked polymerases can be released readily from the halted complexes and engaged in another round of transcription. We have previously reported that about half the RNA fragments of every size synthesized by the nicked polymerase were detected free from immobilized ternary complexes halted by incorporation of 3′-dNMP (15). In contrast, most of the transcripts produced by the intact polymerase were bound to the immobilized complexes.
Acknowledgments
ACKNOWLEDGEMENTS
This work was partially supported by a grant from the Human Genome Project of the Korean Ministry of Science and Technology and by the Brain Korea 21 Project.
References
- 1.Collins F.S., Patrinos,A., Jordan,E., Chakravarti,A., Gesteland,R. and Walters,L. (1998) New goals for the U.S. human genome project: 1998–2003. Science, 282, 682–689. [DOI] [PubMed] [Google Scholar]
- 2.Fu D.-J., Tang,K., Braun,A., Reuter,D., Barnhofer-Memar,B., Little,D.P., O’Donnell,M.J., Cantor,C.R. and Koester,H. (1998) Sequencing exons 5 to 8 of the p53 gene by MALDI-TOF mass spectrometry. Nat. Biotechnol., 16, 381–384. [DOI] [PubMed] [Google Scholar]
- 3.Kirpekar F., Nordhoff,E., Larsen,L.K., Kristiansen,K., Roepstorff,P. and Hillenkamp,F. (1998) DNA sequence analysis by MALDI mass spectrometry. Nucleic Acids Res., 26, 2554–2559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Tang W., Zhu,L. and Smith,L.M. (1997) Controlling DNA fragmentation in MALDI-MS by chemical modification. Anal. Chem., 69, 302–312. [DOI] [PubMed] [Google Scholar]
- 5.Nordhoff E., Cramer,R., Karas,M., Hillenkamp,F., Kirpekar,F., Kristiansen,K. and Roepstorff,P. (1993) Ion stability of nucleic acids in infrared matrix-assisted laser desorption/ionization mass spectrometry. Nucleic Acids Res., 21, 3347–3357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirpekar F., Nordhoff,E., Kristiansen,K., Roepstorff,P., Lezius,A., Hahner,S., Karas,M. and Hillenkamp,F. (1994) Matrix assisted laser desorption/ionization mass spectrometry of enzymatically synthesized RNA up to 150 kDa. Nucleic Acids Res., 22, 3866–3870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Little D.P., Thannhauser,T.W. and McLafferty,F.W. (1995) Verification of 50- to 100- mer DNA and RNA sequences with high-resolution mass spectrometry. Proc. Natl Acad. Sci. USA, 92, 2318–2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tolson D.A. and Nicholson,N.H. (1998) Sequencing RNA by a combination of exonuclease digestion and uridine specific chemical cleavage using MALDI-TOF. Nucleic Acids Res., 26, 446–451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hahner S., Ludemann,H.C., Kirpekar,F., Nordhoff,E., Roepstorff,P., Galla,H.J. and Hillenkamp,F. (1997) Matrix-assisted laser desorption/ionization mass spectrometry (MALDI) of endonuclease digests of RNA. Nucleic Acids Res., 25, 1957–1964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Axelrod V.D. and Kramer,F.R. (1985) Transcription from bacteriophage T7 and SP6 RNA polymerase promoters in the presence of 3′-deoxyribonucleoside 5′-triphosphate chain terminators. Biochemistry, 24, 5716–5723. [DOI] [PubMed] [Google Scholar]
- 11.Nam S.-C. and Kang,C. (1988) Transcription initiation site selection and abortive initiation cycling of phage SP6 RNA polymerase. J. Biol. Chem., 263, 18123–18127. [PubMed] [Google Scholar]
- 12.Sasaki N., Izawa,M., Watahiki,M., Ozawa,K., Tanaka,T., Yoneda,Y., Matsuura,S., Carninci,P., Muramatsu,M., Okazaki,Y. and Hayashizaki,Y. (1998) Identification of stable RNA hairpins causing band compression in transcriptional sequencing and their elimination by use of inosine triphosphate. Proc. Natl Acad. Sci. USA, 95, 3455–3460. [DOI] [PubMed] [Google Scholar]
- 13.Jeong W. and Kang,C. (1997) The histidine-805 in motif of the phage SP6 RNA polymerase is essential for its activity as revealed by random mutagenesis. Biochem. Mol. Biol. Int., 42, 711–716. [DOI] [PubMed] [Google Scholar]
- 14.Butler E.T. and Chamberlin,M.J. (1982) Bacteriophage SP6-specific RNA polymerase. I. Isolation and characterization of the enzyme. J. Biol. Chem., 257, 5772–5778. [PubMed] [Google Scholar]
- 15.Kwon Y.-S. and Kang,C. (1999) Bipartite modular structure of intrinsic, RNA hairpin-independent termination signal for phage RNA polymerase. J. Biol. Chem., 274, 29149–29155. [DOI] [PubMed] [Google Scholar]
- 16.Yoo J. and Kang,C. (2000) Bacteriophage SP6 RNA polymerase mutants with altered termination efficiency and elongation processivity. Biomol. Eng., 16, 191–197. [DOI] [PubMed] [Google Scholar]
- 17.Groderg J. and Dunn,J.J. (1988) OmpT encodes the Escherichia coli outer membrane protease that cleaves T7 RNA polymerase during purification. J. Bacteriol., 170, 1245–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tyagarajan K., Monforte,J.A. and Hearst,J.E. (1991) RNA folding during transcription by T7 RNA polymerase analyzed using the self-cleaving transcript assay. Biochemistry, 30, 10920–10924. [DOI] [PubMed] [Google Scholar]
- 19.Tabor S. and Richardson,C.C. (1990) DNA sequence analysis with a modified bacteriophage T7 DNA polymerase. Effect of pyrophosphorolysis and metal ions. J. Biol. Chem., 265, 8322–8328. [PubMed] [Google Scholar]
- 20.Uptain S.M., Kane,C.M. and Chamberlin,M.J. (1997) Basic mechanisms of transcript elongation and its regulation. Annu. Rev. Biochem., 66, 117–172. [DOI] [PubMed] [Google Scholar]
- 21.Lee J.T., Kim,H., Moon,K., Kim,S. and Kang,C. (1991) Terminator-distal transcribed sequences affecting factor-independent termination efficiency of phage T7 terminator. Mol. Cell., 1, 203–209. [Google Scholar]