Abstract
Fluorescence cell imaging can be used for disease diagnosis and cellular signal transduction. Using a metal nanoshell as molecular imaging agent, we develop a cellular model system to detect CXCR4 chemokine receptor on T-lymphatic cell surface. These metal nanoshells are observed to express enhanced emission intensity and shortened lifetimes due to the near-field interactions. They are covalently bound with anti-CXCR4 monoclonal antibodies for immunoreactions with the target sites of the CXCR4 receptors on the CEM-SS cells. The fluorescence intensity and lifetime cell images are recorded with a time-resolved confocal microscopy. As expected, the emission signals from the metal nanoshells are clearly isolated from the cellular autofluorescence due to strong intensities and distinctive lifetimes. The number of emission spots on the single cell image is estimated by direct count to the emission signals. Analyzing a pool of cell images, a maximal count number is obtained in a range of 200±50. Because there is an average of ∼6000 binding sites on the cell surface, we estimate that one emission spot from the metal nanoshell may represent ∼30 CXCR5 receptors. In addition, the CXCR4 receptors are estimated to distribute on ∼70% area of the cell surface.
Keywords: chemokine receptor 4 (CXCR4), anti-CXCR4 monoclonal antibody (mAb), T-lymphocytic CEM-SS cell, metal nanoshell, Ru (bpy)32+ complex, fluorescence cell image, single molecule count
Introduction
To initiate a HIV infection, the viral envelope glycoprotein gp120 comes in contact with a cellular receptor of T-lymphocyte known as CD4, followed by interaction with co-receptors, e.g., CCR5 or CXCR4.1, 2, 3, 4 As a key co-receptor, CXCR4 thus takes an important role in the HIV-1 infection. In fact, CXCR4 is specifically recognized by the R4 virus during the early phases of the HIV-1 infection.5, 6, 7 Therefore, it is imperative to be able to effectively detect the CXCR4 receptors on the T-cell surface that can serve as an important tool in understanding the molecular mechanism of HIV infection, especially when CXCR4 interacts with its ligand and∕or with viral gp120.
Fluorescence cell imaging shows great promise in disease diagnosis or in elucidating cellular signal transduction.8, 9, 10 In this method, the target molecules on the cell surfaces can be detected by the emission intensity and∕or lifetime. Typically, the molecules of interest on the cell surface are conjugated by molecule imaging agents, which consist of organic fluorophore moieties and targeting functionalities (e.g., antibodies, peptides, or special ligands, through immunoreactions).11, 12 However, cell imaging is often problematic due to interference from cellular autofluorescence and intrinsic drawbacks from the conventional organic fluorophores in cell imaging. Cellular autofluorescence is fluorescence from the substrate other than the fluorophore of interest in cell imaging. Autofluorescence arises principally from the mitochondria and liposome of the cell. It is a type of broad spectrum from UV to infrared wavelength but has a window in the near-infrared region. Thus, it is almost impossible to distinguish the emission of fluorophores from cellular autofluorescence by spectroscopy in cell imaging. On the other hand, organic fluorophores often display weak emission intensity, rapid photobleaching, and strong photoblinking13 which may influence the observations to the target molecules. Most importantly, the lifetime of organic fluorophores mostly ranges from 2–10 ns, close to that of cellular autofluorescence, which leads to difficulties in the isolations of the probe molecules from the cellular background in lifetime-resolved imaging. As a result, the target molecules on the cell surfaces cannot be accurately estimated. Hence, it is essential to develop novel imaging agents that can improve the detectability and accuracy of the target molecules in cell imaging.14
During the past decade, metal nanoparticles with fluorescence were being developed.15, 16, 17, 18, 19, 20, 21 They are designed in two configurations: metal nanosphere and metal nanoshell. To the metal nanosphere, the fluorophores are bound on the metal surface. To the metal nanoshell, the fluorophores are encapsulated in the core. We are particularly interested in metal nanoshells and use them as molecular imaging agents to detect the single CXCR4 molecules on the cell surface by the imaging method in this research. To the metal nanoshell, although part of light-induced electromagnetic waves must be screened out to the far field, according to the calculations from other groups,22, 23, 24, 25 the majority of the energy is left in the interior space of the metal shell and uniformly distributed in the core area. As a result, the fluorophores throughout the core can be coupled equally and efficiently with the interior uniform field, leading to greatly enhanced fluorescence.26, 27, 28 Importantly, the near-field interaction of the fluorophore with the metal nanostructure can increase the radiative rate of a fluorophore, which may increase the intrinsic decay rate of fluorophores, leading to a dramatic reduction in lifetime.18 Therefore, these metal nanoshells can be developed as the time-resolved fluorescent probes in the lifetime cell imaging. In addition, the near-field interactions can extend photobleaching and reduce photoblinking.17 The versatile chemistry on the metal surfaces may promise binding of the probe moieties of biological interest to the metal nanoshells.29 In fact, the fluorescent metal nanoparticles have been used as molecule imaging agents for the detection of target molecules on the cell surfaces19, 20 or in the cells.21
In this paper, the metal nanoshells were designed to have high brightness and a distinctive lifetime relative to cellular autofluorescence and used as molecular imaging agents to detect the CXCR4 receptors on the T-lymphocytic cell surface. Anti-CXCR4 mAb molecules were covalently bound on the metal nanoshells for immuno-reactions with the CXCR4 receptors on the CEM-SS cells that were derived from CD4-positive T-lymphocytes. The emission intensity and lifetime cell images were recorded at the single cell level with a time-resolved confocal microscope. Based on the count numbers and spatial patterns of emission signals from the metal nanoshells on the cell images, the CXCR4 receptors were evaluated in terms of the expression numbers and mapping distributions on the CEM-SS cell surfaces. Because CXCR4 is a key binding co-receptor by viral gp120 during the HIV infection, the results may provide a reference to our future understanding of the HIV infection on the cell membrane of T-lymphocytes.
Experimental Procedures
All reagents and spectroscopic grade solvents were used as received from Sigma-Aldrich. Nanopure water (>18.0 MΩ.cm) purified by Millipore Milli-Q gradient system was used in the experiments. Anti-CXCR4 monoclonal antibody was obtained from the NIH AIDS Research and Reference Reagent Program. Rhodamine labeled anti-goat IgG were from VWR. RC dialysis membrane (MWCO 50,000) was obtained from Spectrum Laboratories, Inc.
Preparation of Anti-CXCR4 mAb-Ag Nanoshell Complex
In this report, the metal nanoshells were prepared with the 50 nm silica cores∕10 nm thick silver walls as previous report.21 The complexes were encapsulated in the silica cores with the loading number of approximately 120.21 These metal nanoshells were dispersed in water with 10 mM hexa(ethyleneglycol)mono-11-(acetylthio)undecyl ether for 2 h, which led to the assembling of organic monolayers on the metal surfaces. Subsequently, they were partially substituted by 11-mercapto-undecanoic acid via ligand exchanges in order to bind with anti-CXCR4 monoclonal antibdies (mAbs).29, 30 Typically, the silver nanoshells (1×10−8 M) and 11-mercapto-undecanoic acid (1×10−6 M) are codissolved in a mixture solvent of ethanol and water (v∕v = 1∕1). The solution was stirred for 24 h at room temperature. The suspension was removed by centrifugation, and the residue was rinsed with ethanol and water. The recovered metal nanoshells (1×10−8 M) were codissolved with anti-CXCR4 mAb (1×10−6 M) in 10 mM phosphate buffered saline (PBS) buffer solution followed by addition of 1-ethyl-3-(3-dimethylaminopropyl)-carbodiimide hydrochloride (2×10−5 M) in excess amount. The solution was stirred continuously for 2 h at room temperature. The suspension was removed by centrifugation at 6000 rpm, and the residue was rinsed with 10 mM PBS buffer. After dialysis against 10 mM PBS buffer solution, the mAb-Ag nanoshell complexes were redispersed in 10 mM PBS buffer solution for the cell incubations.
In the same strategy, the goat anti-IgG mAb molecules were also covalently bound on the metal nanoshells to use as negative controls for the specific immunointeractions with the target CXCR4 receptors on the cell surfaces.
Culturing and Labeling CEM-SS Cells with CXCR4 mAb-Ag Nanoshell Complex
The CEM-SS cell line, a derivative of CXCR4(+) T-lymphocytes, was grown in RPMI-1640 culture medium (Sigma, St. Louis, MO) supplemented with 10% (v∕v) heat-inactivated fetal bovine serum (Atlanta Biologicals Inc., Lawrenceville, Georgia) and contained 200 units∕ml penicillin, 200 units∕ml streptomycin (Invitrogene, Carlsbad, CA), and recombinant human interleukin (100U∕ml) (Roche, Indianapolis, Indiana) for six days prior to the conjugation experiments.31 The number of cells was counted to be ca. 5×106 cells∕mL. The CEM-SS cells in 500 μL aliquots were incubated with the mAb-Ag nanoshell complexes in the different concentrations of 1, 3, 10, 20, 50, 100, 200, and 400 × 10−12 M (pM) at room temperature. The incubations took 2 h. After being washed with PBS-Mg solution, the cell lines were resuspended in 500 μL of 10 mM PBS buffer solution, and 20 μL cell-suspended solutions were cast on the cleaned glass coverslips and dried in air for cell imaging.
Spectral, Imaging, and Transmission Electron Micrograph Measurements
Absorption spectra were determined on a Hewlett Packard 8453 spectrophotometer. Ensemble fluorescence spectra were performed on a Cary Eclipse Fluorescence Spectrophotometer. For the transmission electron micrograph measurements, the nanoparticle samples, including the silica nanosphere and metal nanoshells, were diluted to nanomolar scale in ethanol and cast on the copper grids (200 mesh) with standard carbon-coated Formvar films (200–300 Å). The samples were dried in air. The images were taken with a side-entry Philips electron microscope at 120 keV. The size distributions were analyzed with Scion Image Beta Release 2 on the base on at least 100 images.
Fluorescence cell imaging was performed on a time-resolved scanning confocal microscopy (MicroTime 200, PicoQuant), which consists of an inverted confocal microscope coupled to a high-sensitivity detection setup. A single-mode pulsed laser diode (470 nm, 100 ps, 10 MHz) was used as the excitation source. An oil immersion objective (Olympus, 100×, 1.3 NA) was used for focusing the laser light onto the sample and collecting the emission signal. The emission passed a dichroic mirror, focused onto a 75-μm pinhole for a spatial filtering to reject out-of-focus signals, and recorded on a single-photon avalanche diode (SPCM-AQR-14, Perkin Elmer Inc.). Bandpass filters were used to eliminate the excitation residual. The data were collected with a TimeHarp 200 board and stored in the time-tagged time-resolved mode that allows every detected photon with its individual timing and detection channel information.
Results and Discussion
In this research, according to the method in our previous report,21 the metal nanoshells were generated with 50-nm silica cores, 10-nm metal walls, and ∼120 complexes in one core. The metal nanoshells were coated by the monolayers of hexa(ethyleneglycol)mono-11-(acetylthio)undecyl ether so they displayed good chemical stability in buffer solution. The metal plasmon resonance displayed a broadened absorbance with a maximum at ∼400 nm. On an excitation at 450 nm, the emission displayed a maximal at 605 nm. These results were in accordance to the observations in our previous report.26, 27, 28
The organic monolayers on the metal nanoshells were partially substituted by the terminal-carboxylate ligands via ligand exchange.30 Then, anti-CXCR4 mAb molecules were covalently bound on the metal nanoshell surfaces via condensation.19, 20 We could not provide direct evidence to prove the occurrences of these surface reactions. Instead, the Rhodamine 6G-labeled antigoat IgG mAbs were treated in the same strategy with the metal nanoshells without the complex encapsulations. On excitation at 510 nm, the anti-IgG mAb-Ag complexes exhibited a maximal emission from Rhodamine 6G at 551 nm, demonstrating the IgG mAb molecules were indeed bound on the metal nanoshells. Thus, the anti-CXCR4 mAb molecules were believed to be able to bind on the metal nanoshells. We also tested the number of mAb molecules on each metal nanoshell might be an important influence to its immunoreaction capability with the target site on the cell.32 The IgG mAb-metal nanoshell complexes were treated with NaCN to dissolve the metal and release the Rhodamine 6G-labeled anti-IgG mAbs.33 After adding several drops of 0.1 M NaCN aqueous solution, the color of the plasmon resonance of the solution was observed to progressively disappear in correspondence to the dissolution of metal walls on the silica cores. The concentration of metal nanoshell in buffer solution was estimated from the absorbance intensity of plasmon resonance before treatment, and the concentration of released IgG mAb was determined from the emission intensity of released mAb after treatment. Through a molar ratio of IgG mAb over the metal nanoshell, the number was estimated to be 17.3. As discussed above, the anti-CXCR4 mAbs were regarded to load on the nanoshells with the same number.
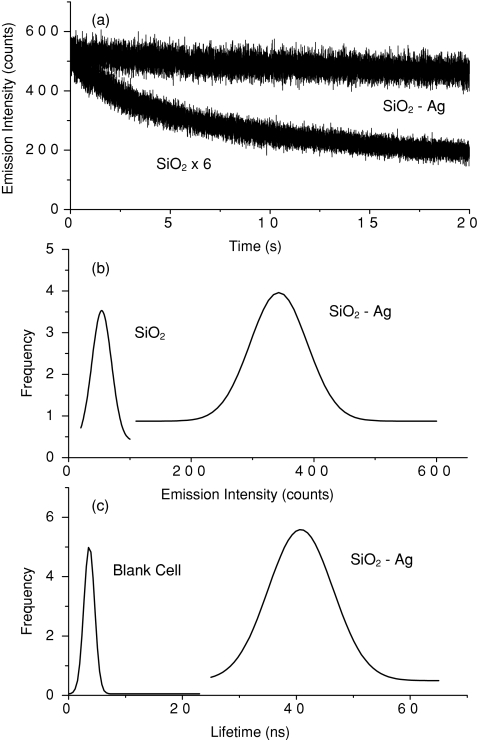
With a scanning confocal microscopy, we tested the intrinsic optical properties of single anti-CXCR4 mAb-Ag nanoshell complexes. The time trace of a single complex was observed to express a direct decay of emission intensity [Fig. 1a], representing the presence of multiple fluorophores on a single complex. Similarly, the time trace of a single metal-free silica sphere also expressed a direct decay but a much lower intensity [Fig. 1a]. A minimum of 50 fluorescent spots from the single complexes as well as the silica spheres were collected for the analyses of emission intensity [Fig. 1b] and lifetime [Fig. 1c]. Although the lifetimes of silica spheres without metal were too long to be accurately detected by the current microscopy, the histogram results clearly revealed that the emission intensity was enhanced up to sixfold and the lifetime was reduced dramatically to 45 ns by the metal shells. The changes of optical properties were caused by near-field interactions of fluorophores with the metal.18 We also note that although shortened by the metal, the lifetime of the metal nanoshell was still significantly longer than the lifetime of cellular autofluorescence (2–10 ns). Thus, the emission signals from metal nanoshells are expected to be isolated from the cellular backgrounds on lifetime cell imaging. The quantum yields of complex in water and in the silica gel are reported to be 0.04 and 0.11,34 respectively. We did not measure the value of the metal nanoshells in the current measurement. But according to enhanced emission intensity by the metal nanoshells, we expect that the quantum yield of the metal nanoshells to be ∼0.66.
Figure 1.
(a) Time-trace profiles of a typical single silica sphere without metal and a typical single metal nanoshell. (b,c) respectively present the histograms of emission intensities from the single silica spheres and metal nanoshells and the lifetimes from metal nanoshells. In (c), the distribution of lifetime throughout the blank cell is also presented.
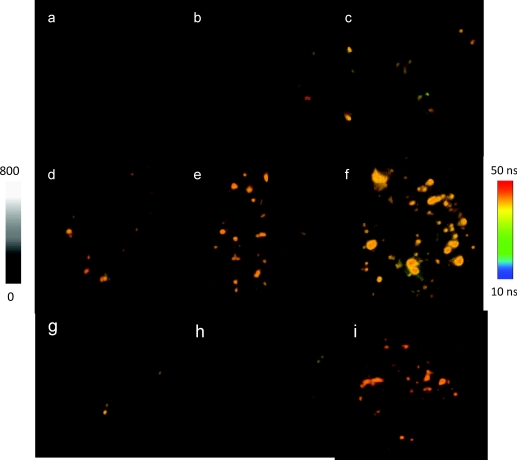
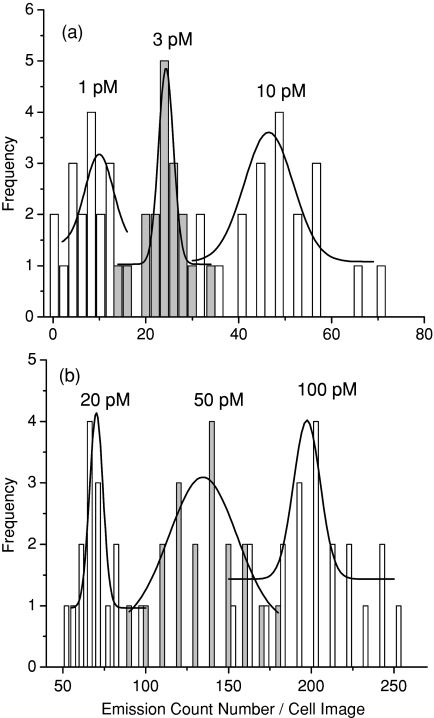
By incubation, the anti-CXCR4 mAb-Ag nanoshell complexes were immunointeracted with the target sites on the CEM-SS cells. To observe the immunoreactions, the incubations were carried out with an increase of anti-CXCR4 mAb-Ag complex concentration, ranging from 1×10−12 to 400 × 10−12 M in incubation solution. Representative emission intensity and lifetime cell images were shown in Fig. 2. Almost no emission spot from the complex was observed out of the collected cell images, indicating the complexes were properly conjugated with the target sites on the cell surfaces. With strong emission intensity and distinctive lifetime, the emission spots from the complexes were clearly identified from cellular autofluorescence. The amount of emission spots on the single cell images was dependent on the concentration of the complex in the incubation solution. At a low conjugation, the emission signals were isolated as small and bright spots, and the numbers were able to be accurately counted. By increasing the conjugation amount, the emission signals from the complexes became dense and continuous on the cell images, which resulted in the discrete areas with a strong intensity and long lifetime, implicating that the complexes were conjugated as clusters on the cell surfaces.
Figure 2.
Representative fluorescent intensity and lifetime cell images of CEM-SS cells incubated in solution with anti-CXCR4 mAb-Ag complex at (a) 0, (b) 1, (c) 3, (d) 10, (e) 50, and (f) 100 pM, respectively. (g,h) Images were of the negative controls of CEM-SS cells that were incubated in solution with the anti-goat IgG-Ag nanoshell complex at (g) 10 and (h) 100 pM. (i) Image on which the lifetimes of <10 ns were removed. Scale of the diagrams was 20 × 20 μm, and the resolution of diagrams was 400 × 400 pixel with an integration of 0.6 ms∕pixel.
With the same strategy, the CEM-SS cells were also incubated with anti-IgG mAb-Ag complexes. The cell images were collected as negative controls of immunoreactions [Figs. 2g, 2h]. Only seldom were emission signals from the complexes shown on the cell images, and the numbers were also absent of a significant concentration-dependence, which further confirms that anti-CXCR4 mAb-Ag complexes were principally conjugated with the target sites of CXCR4 receptors on the cell surfaces.
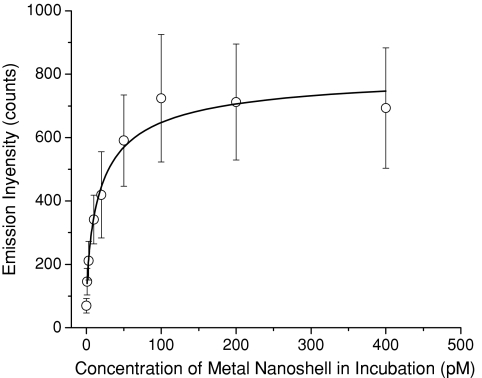
To relate the intrinsic optical properties over a cell image with the number of target molecules on the cell surface, we analyzed the emission intensity and lifetime throughout the entire cell image using PicoQuant analysis software. At each incubation concentration, a minimum of 20 cell images were treated to achieve the average values. The obtained average emission intensities were plotted against the concentrations of anti-CXCR4 mAb-Ag complexes in incubation solution (Fig. 3). The curve exhibited an increase of intensity with the concentration and reached saturation at 100 pM. The saturation fully represents conjugations of complexes with the target sites on the cell surfaces.
Figure 3.
Dependence of the average emission intensity throughout the entire cell image on the metal nanoshell concentration in incubation solution.
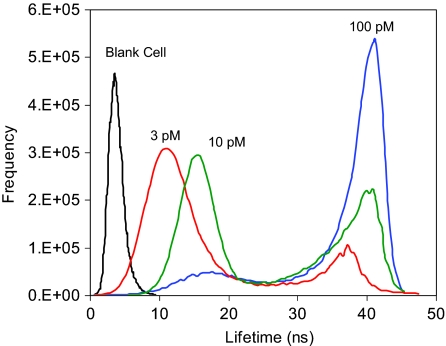
In some cases, lifetime is regarded as a more important factor in cell imaging.13 In this paper, for the mock immunoreacted “blank” cell, the lifetime distribution throughout the entire cell image displayed a maximum at 3 ns (Fig. 4). This value was regarded from cellular autofluorescence of the cell line. By increasing the concentration of complex in incubation solution, the lifetime cell image displayed a significant rise at 40 ns, accompanied by a simultaneous decrease of the shorter-lifetime component. The longer-lifetime component was regarded from the conjugated complexes, whereas the shorter-lifetime component was from cellular autofluorescence. The longer- and shorter-lifetime components were not significantly overlapped corresponding to clear isolations of the emission signals of mAb-Ag complexes from cellular autofluorescence on the cell image. In addition, the shorter-lifetime component was observed to progressively shift to longer with an increase of conjugation amount. As with the emission intensity, the average lifetime over the entire cell image also expressed an increase with the incubation concentration and reached saturation at 100 pM, indicating complete conjugation of the complexes on the cell surface.
Figure 4.
Lifetime distributions throughout the entire cell images that were collected from cells incubated with different metal nanoshell concentrations including 0, 3, 10, and 100 pM.
In order to isolate the emission spots of mAb-Ag complexes for precise counting, lifetimes of <10 ns (mostly from cellular autofluorescence) were removed using OriginPro-7 software. Although counting of the emission spots on a single cell image was straightforward, the actual number was approximate because instead of individual spots, some emission signals were shown as continuous clusters on the cell image under high conjugations. In the actual treatments, the large clusters were divided into small ones to make their sizes approximately equivalent to that of a single spot. The count numbers obtained in this method showed a significant increase with the incubation concentration of complex (Fig. 5), and the saturation number was approximately 200±50 at 100 pM. It is conceivable that this number does not reflect the actual amount of CXCR4 site on the cell surface. In fact, an early report shows that the CEM-SS cell has the expression of CXCR4 in the order of 6 × 103.35 If it is real, then the count number estimated in this research seems drastically undercounted. In the other words, we can infer that one emission spot on the cell image may represent about 30 actual CXCR4 receptors. This observation is interpreted with two possible reasons. The first is the resolution of confocal microscope. It is known that the x-y–dimensional resolution of the confocal microscope is 200 nm. However, the size of the complex is ∼70 nm. Thus, one emission spot on the cell image is supposed to be able to contain the signals from several complexes that cannot be resolved. The maximum is ∼9. The second is the presence of multiple anti-CXCR4 mAbs on a single complex. The loading number is 17. Thus, a single complex is supposed to be able to immunoreact with multiple CXCR4 receptors on the cell surface up to 17. Of course, this number cannot be that high when considering the three-dimensional structure and immunoreaction capability of a complex on the cell surface. But combining these two factors, the estimation number of 30 is approximately acceptable in this research.
Figure 5.
The count numbers of emission spots on the single cell images that were estimated from cells incubated with different concentrations.
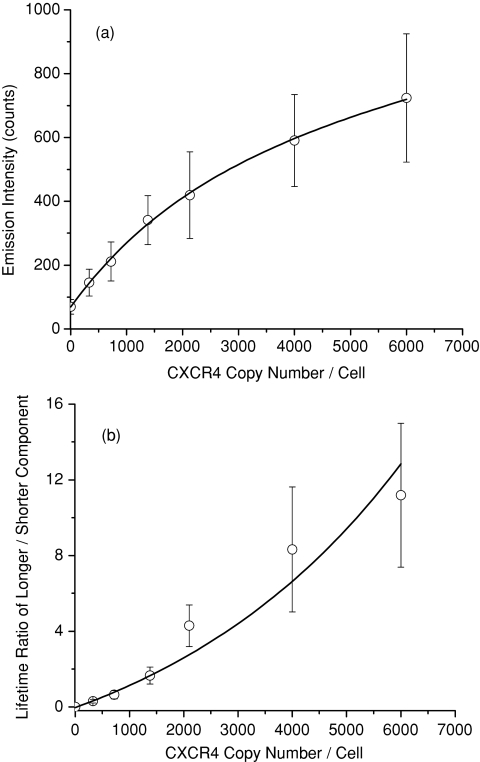
Assuming that the estimation is close to the reality, we converted the count numbers of complexes on the cell images to the actual amount of CXCR4 receptors on the cell surfaces by predicting 30. The average emission intensity and lifetime over the cell image were restively plotted against the estimated CXCR4 numbers ]Figs. 6a, 6b]. To the lifetime figure, the ratio of longer-component over shorter-component was used to express its typical change with the estimated amount on the cell surface. Both emission intensity and lifetime parameters were increased with the number. It must be emphasized herein that the two curves are only used to illustrate the nature of the CXCR4 amount on the cell surface with the optical properties on the cell image and not derive the actual number of CXCR4 receptors.
Figure 6.
Dependences of the (a) emission intensity and (b) lifetime on the estimated number of CXCR4 receptors on the single cell surface.
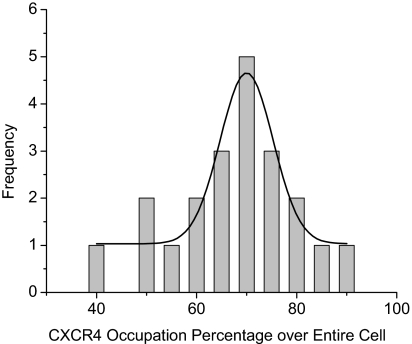
Unlike flow cytometry, the cell imaging developed in this paper may provide a direct observation to the spatial distribution of target molecules on a cell surface. For instance, the CXCR4 receptors were observed to distribute relatively evenly throughout the cell surface. In addition, the coverage percentage can be used to imply the occupation of target molecules over the entire cell surface. It was estimated on the basis of lifetime cell images. Typically, the lifetimes on a single cell image were entirely set to be 1 using the OriginPro-7 software followed by integration. The integrated value may represent the entire area of the cell. Subsequently, the lifetimes of <10 ns were removed and the residual lifetimes were set to be 1 followed by integration. This integrated value may represent the occupation area of CXCR4 receptors on the cell surface. The ratio of two values thus is regarded as the coverage percentage of CXCR4 receptors throughout the entire cell surface. A minimum of 20 cell images were analyzed to obtain the histogram (Fig. 7). A maximum is obtained at ∼0.7, indicating that the CXCR4 receptors are distributed on a 70% area of the total cell surface.
Figure 7.
Histogram of coverage percentage of the CXCR4 receptors throughout the cell surface.
Conclusion
In this paper, we used metal nanoshells as molecular imaging agents to detect the CXCR4 co-receptors on the T-lymphatic cell surface. Metal nanoshells were observed to display strong emission intensity and distinctive lifetime from cellular autofluorescence that might allow clear isolations of the molecules of interest (i.e., CXCR4) from the cellular background on the cell images. It is important to note that, although we have provided the results of a proof-to-concept study demonstrating a potentially rapid and simple method for the quantification of target molecules on the cell surface, with further analytical and clinical validations, we expect this approach may provide a simple and rapid method of detecting the CXCR4 receptor on the surface of T-cells, which is biologically important in an HIV infection during the interaction of viral gp120 with the receptors and co-receptors.
Acknowledgment
The authors thank the NIH AIDS Research and Reference Reagent Program for providing the anti-CXCR4 mAbs. This research was supported in part by grants from NIH (EB009509, HG-002655, HG005090, EB006521, and CA134386).
References
- Levine B. and Sodora D. L., “HIV and CXCR4 in a kiss of autophagic death,” J. Clin. Invest. 116, 2078–2080 (2006). 10.1172/JCI29447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baleux F., Loureiro-Morais L., Hersant Y., Clayette P., Arenzana-Seisdedos F., Bonnaffé D., and Lortat-Jacob H. A., “Synthetic CD4-heparan sulfate glycoconjugate inhibits CCR5 and CXCR4 HIV-1 attachment and entry,” Nat. Chem. Biol. 5, 743–748 (2009). 10.1038/nchembio.207 [DOI] [PubMed] [Google Scholar]
- Grivel J.-C., and Margolis L. B., “CCR5- and CXCR4-tropic HIV-1 are equally cytopathic for their T-cell targets in human lymphoid tissue,” Nat. Med. 5, 344–346 (1999). 10.1038/6565 [DOI] [PubMed] [Google Scholar]
- Dragic T., et al. , “HIV-1 entry into CD4(+) cells is mediated by the chemokine receptor CC-CKR-5,” Nature 381, 667–673 (1996). 10.1038/381667a0 [DOI] [PubMed] [Google Scholar]
- Tchou I., Misery L., Sabido O., Dezutter-Dambuyant C., Bourlet T., Moja P., Hamzeh H., Peguet-Navarro J., Schmitt D., and Genin C., “Functional HIV CXCR4 coreceptor on human epithelial Langerhans cells and infection by HIV strain X4.” J. Leukocyte Biol. 70, 313–321 (2001). [PubMed] [Google Scholar]
- Bacon K. et al. , “Chemokine∕chemokine receptor nomenclature,” J. Interferon Cytokine Res. 22, 1067–1068 (2002). 10.1089/107999002760624305 [DOI] [PubMed] [Google Scholar]
- Agrawal L., Qingwen J., VanHorn-Ali Z., Nicolescu I. V., McDermott D. H., and Murphy P. M., “Alkhatib G role for CCR5 Delta 32 protein in resistance to R5, R5X4, and X4 human immunodeficiency virus type 1 in primary CD4(+) cells,” J. Virol. 78, 2277–2282 (2004). 10.1128/JVI.78.5.2277-2287.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Licha K., Riefke B., Ntziachristos V., Becker A., Chance B., and Semmler W., “Hydrophilic cyanine dyes as contrast agents for near-infrared tumor imaging: synthesis, photophysical properties and spectroscopic in vivo characterization,” Photochem. Photobiol. 72, 392–398 (2000). [DOI] [PubMed] [Google Scholar]
- Mahmood U. and Weissleder R., “Near-infrared optical imaging of proteases in cancer,” Mol. Cancer Therapeut. 2, 489–496 (2003). [PubMed] [Google Scholar]
- Nakanishi J., Nakajima T., Sato M., Ozawa T., Tohda K., and Umezawa Y., “Imaging of conformational changes of proteins with a new environment-sensitive fluorescent probe designed for site-specific labeling of recombinant proteins in live cells,” Anal. Chem. 73, 2920–2938 (2001). 10.1021/ac001528p [DOI] [PubMed] [Google Scholar]
- Ntziachristos V. and Chance B., “Probing physiology and molecular function using optical imaging: applications to breast cancer,” Breast Cancer Res. 3, 41–46 (2001). 10.1186/bcr269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissleder R., Tung C. H., Mahmood U., and Bogdanov A., “In vivo imaging of tumors with protease-activated near-infrared fluorescent probes,” Nat. Biotechnol. 17, 375–378 (1999). 10.1038/7933 [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., Principles of Fluorescence Spectroscopy, 3rd ed., Springer, New York: (2006). [Google Scholar]
- Kamat P. V., “Photophysical, photochemical and photocatalytic aspects of metal nanoparticles,” J. Phys. Chem. B 106, 7729–7744 (2002). 10.1021/jp0209289 [DOI] [Google Scholar]
- Jain P. K., Huang X., El-Sayed I. H., and El-Sayed M. A., “Noble metals on the nanoscale: optical and photothermal properties and some applications in imaging, sensing, biology, and medicine,” Acc. Chem. Res. 41, 1578–1586 (2008). 10.1021/ar7002804 [DOI] [PubMed] [Google Scholar]
- Bardhan R., Grady N. K., Cole J. R., Joshi A., and Halas N. J., “Fluorescence enhancement by Au nanostructures: nanoshells and nanorods,” ACS Nano. 3, 744–752 (2009). 10.1021/nn900001q [DOI] [PubMed] [Google Scholar]
- Lakowicz J. R., “Radiative decay engineering 5: metal-enhanced fluorescence and plasmon emission,” Anal. Biochem. 337, 171–194 (2005). 10.1016/j.ab.2004.11.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J. R., “Radiative decay engineering: biophysical and biomedical applications,” Anal. Biochem. 298, 1–24 (2001). 10.1006/abio.2001.5377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fu Y., Liang D., Nowaczyk K., Zhao R. Y., and Lakowicz J. R., “Single cell fluorescence imaging using metal plasmon-coupled probe 2: lifetime image and single molecule counting,” Nano Lett. 8, 1179–1186 (2008). 10.1021/nl080093z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fu Y., Liang D., Zhao R. Y., and Lakowicz J. R., “Fluorescent avidin-bound silver particle: a strategy for single target molecule detection on a cell membrane,” Anal. Chem. 81, 883–889 (2009). 10.1021/ac801932m [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fu Y., Mei Y., Jiang F., and Lakowicz J. R., “Fluorescent metal nanoshell probe to detect single miRNA in lung cancer cell,” Anal. Chem. 82, 4464–4471 (2010). 10.1021/ac100241f [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enderlein J., “Fluorescence enhancement of a single dye within a spherical metallic nanocavity,” Appl. Phys. Lett. 80, 315–317 (2002). 10.1063/1.1434314 [DOI] [Google Scholar]
- Lassiter J., Knight M. W., Mirin N. A., and Halas N. J., “Reshaping the plasmonic properties of an individual nanoparticle,” Nano Lett. 9, 4326–4332 (2009). 10.1021/nl9025665 [DOI] [PubMed] [Google Scholar]
- Hao E., Li S., Bailey R. C., Zou S., Schatz G. C., and Hupp J. T., “Optical properties of metal nanoshells,” J. Phys. Chem. B 108, 1224–1229 (2004). 10.1021/jp036301n [DOI] [Google Scholar]
- Olson T. Y., Schwartzberg A. M., Orme C. A., Talley C. E., O’Connell B., and Zhang J. Z., “Hollow gold-silver double-shell nanospheres: structure, optical absorption, and surface-enhanced Raman scattering,” J. Phys. Chem. C 112, 6319–6329 (2008). 10.1021/jp7116714 [DOI] [Google Scholar]
- Zhang J., Fu Y., and Lakowicz J. R., “Emission behavior of complex in silver nanoshell–enhanced self-quenching by metal nanostructure,” J. Phys. Chem. C 111, 1955–1961 (2007). 10.1021/jp063996u [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Gryczynski I., Gryczynski Z., and Lakowicz J. R., “Dye-labeled silver nanoshell—bright particle” J. Phys. Chem. B 110, 8986–8991 (2006). 10.1021/jp057032z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Fu Y., and Lakowicz J. R., “Luminescent silica core∕silver shell encapsulated with Eu(III) complex,” J. Phys. Chem. C 113, 19404–19410 (2009). 10.1021/jp906742q [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daniel M.-C. and Astruc D., “Gold nanoparticles: assembly, supramolecular chemistry, quantum-size-related properties, and applications toward biology, catalysis, and nanotechnology,” Chem. Rev. 104, 293–346 (2004). 10.1021/cr030698 [DOI] [PubMed] [Google Scholar]
- Templeton A. C., Wuelfing W. P., and Murray R. W., “Monolayer-protected cluster molecules,” Acc. Chem. Res. 33, 27–36 (2000). 10.1021/ar9602664 [DOI] [PubMed] [Google Scholar]
- Yuan F., Dellian M., Fukumura D., Leunig M., Berk D. A., Torchilin V. P., and Jain R. K., “Vascular permeability in a human tumor xenograft: molecular size dependence and cutoff size,” Cancer Res. 55, 3752–3756 (1995). [PubMed] [Google Scholar]
- Fang, X., and Tan, W., “Aptamers generated from cell-SELEX for molecular medicine: a chemical biology approach,” Acc. Chem. 43, 48–57 (2010). 10.1021/ar900101s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosi N. L. and Mirkin C. A., “Nanostructures in biodiagnostics,” Chem. Rev. 105, 1547–1562 (2005). 10.1021/cr030067f [DOI] [PubMed] [Google Scholar]
- Huang X., Kovaleski J. M., and Wirth M. J., “Spectroscopic probing of mixed-mode adsorption of to silica,” Anal. Chem. 68, 4119–4123 (1996). 10.1021/ac960587t [DOI] [PubMed] [Google Scholar]
- Lee B., Sharron M., Montaner L. J., Weissman D., and Doms R. W., “Quantification of CD4, CCR5, and CXCR4 levels on lymphocyte subsets, dendritic cells, and differentially conditioned monocyte-derived macrophages,” Proc. Natl. Acad. Sci. USA 96, 5215–5220 (1999). 10.1073/pnas.96.9.5215 [DOI] [PMC free article] [PubMed] [Google Scholar]