Abstract
Signal transduction can be defined as the coordinated relay of messages derived from extracellular cues to intracellular effectors. More simply put, information received on the cell surface is processed across the plasma membrane and transmitted to intracellular targets. This requires that the activators, effectors, enzymes, and substrates that respond to cellular signals come together when they need to.
The quest to discover the full complement of cell-signaling components has achieved notable success, and so the next challenge is to establish how these pieces work in concert. In solving this cellular jigsaw puzzle, it is evident that sophisticated regulatory mechanisms ensure that signaling enzymes encounter their intracellular substrates in the right place and at the right time. This requires a delicate balance between two apparently opposing processes: the diffusion of information through the cytoplasm and nucleus, and the processing of information by immobilized multiprotein complexes. Cells have evolved a variety of clever ways to fulfill these requirements: (i) signal-dependent formation of protein complexes; (ii) processing of signals through preassembled multiprotein complexes; (iii) enzyme regulation by subcellular localization; and (iv) temporal control of signaling pathways. Here we highlight recent advances in our understanding of these essential regulatory processes.
Signal-Dependent Formation of Protein Complexes
Information relay from one cellular location to another often requires the dynamic formation of protein complexes. This can be initiated by posttranslational modification, switchlike functions of guanosine triphosphatases (GTPases), or protein oligomerization to generate pockets of concentrated enzyme activity. Signaling proteins typically have a modular organization, being composed of domains with binding or catalytic functions, interspersed with regions that serve as docking or substrate sites for other molecules. Currently, about 100 specialized protein-interaction modules have been identified that recognize a plethora of chemical signals. This section will compare and contrast signaling pathways that use phosphorylation, phosphoinositides, ubiquitination, and acetylation as their primary means of communication.
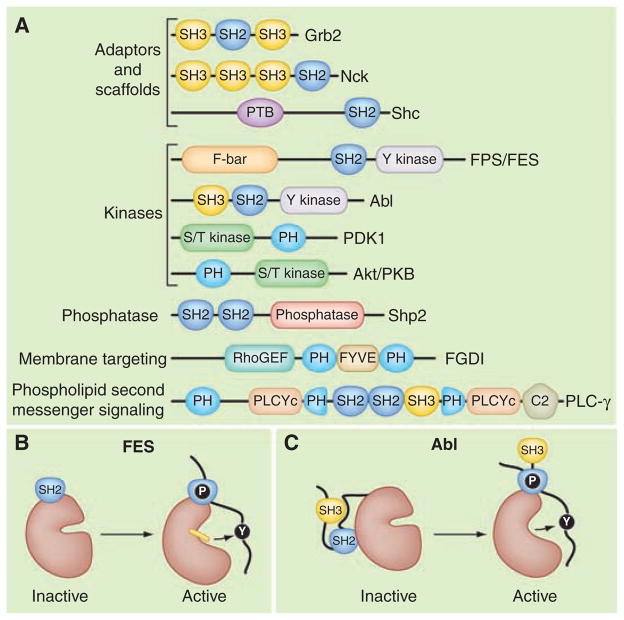
The Src homology 2 (SH2) domain is the archetypal protein-interaction module. Initially discovered in the P130Gag-Fps oncoprotein, SH2 domains consist of about 100 amino acids that bind to specific phosphotyrosine (pY)–containing peptide motifs (1, 2). The human genome encodes 120 SH2 domains embedded in a variety of proteins (Fig. 1A). Most SH2 domain–containing proteins are recruited to sites of tyrosine phosphorylation to aid in the construction of molecular machines (3). The molecular glue in these transduction units is frequently a cohort of adaptor proteins (Fig. 1A). These noncatalytic organizing proteins contain a domain that selectively recognizes the activated receptor (an SH2 domain in the case of receptor tyrosine kinases), linked to domains such as SH3, that engage specific downstream targets, typically by binding to proline-rich sequences. Multivalent modular proteins such as Grb2 (SH3-SH2-SH3) and Nck (SH3-SH3-SH3-SH2) exemplify this strategy (Fig. 1A). For example, growth factor–induced autophosphorylation of receptor tyrosine kinases on the inner face of the plasma membrane creates pockets of phosphotyrosine. This recruits the Grb2 adaptor protein and effector proteins such as phosphatidylinositol 3-kinase (PI3K) or phospholipase–Cγ (PLC-γ) to initiate downstream signaling pathways that contribute to oncogenesis and cancer cell proliferation (4). Accordingly, if the autophosphorylation of receptor tyrosine kinases is suppressed pharmacologically, the assembly of the downstream signaling complexes may be halted, providing a chance to slow the progression of certain cancers (5). Despite their apparent molecular simplicity, adaptor proteins such as Nck, which links pY signals to the actin cytoskeleton, can influence sophisticated biological processes. In neurons, Nck proteins are required for the guidance of spinal cord axons, and the formation of neuronal circuits required for walking (6). Nck is also required for the proper architectural organization of podocytes (specialized cells forming a filtration barrier in the kidney) and is a candidate for involvement in human diabetic nephropathy (7).
Fig. 1.
The modular organization of signaling proteins. (A) Schematic diagram depicting the modular domain organization of selected adaptor and scaffolding proteins (Grb2, Nck, Shc); the tyrosine kinases (Fps/FES and Abl); the serine/threonine (S/T) kinases (PDK 1 and Akt/PKB); the protein tyrosine phosphatase (Shp2); a membrane-targeted Rho guanine nucleotide exchange factor (FGDI); and phospholipase C–gamma (PLC-γ). Individual protein modules are indicated: Src homology 2 (SH2); Src homology 3 (SH3); phosphotyrosine binding (PTB); FCHo2-Bin/Amphiphysin/Rvs domain (F-BAR); Fab-1, YGL023, Vps27, and EEA1 domain (FYVE); and Ca2+− dependent membrane-targeting (C2) module. Enzymatic units in each protein are named. (B) Model depicting the active conformation of FES where the SH2 and tyrosine kinase domains form a single functional unit bound to a primed substrate (8). (C) Abl is maintained in an inactive state through the docking of the SH2 domain on the back face of the catalytic core. These intramolecular interactions are broken upon substrate binding (11).
SH2 domains can also directly influence enzymatic activity (Fig. 1, B and C). For example, in the active configuration of the human FES cytoplasmic tyrosine kinase, the SH2 domain is integrated with the kinase domain to form a single functional unit that is only fully active when bound to a primed substrate (8). This nicely illustrates that the FES SH2 domain is a multifunctional device with a conventional phosphopeptide-binding site and an entirely distinct surface that stabilizes the active kinase [Fig. 1B (8)]. In other molecular contexts, SH2 domains suppress tyrosine kinase activity. When fused to SH3 domains, they inhibit enzymes such as Abl, Src, Lyn, and Fyn. Docking of the SH2 domain on the back face of the catalytic core allows a flexible linker to form an internal binding site for the SH3 domain (Fig. 1C). Consequently, this SH2-SH3 unit stabilizes an inactive conformation of the enzyme (9, 10). Furthermore, the orientation of each SH2-SH3 unit subtly, but distinctly, shapes the topology of the substrate and adenosine 5′-triphosphate (ATP)–binding pockets in each enzyme. This latter feature may help explain the selectivity of certain ATP inhibitor drugs such as imatinib (Gleevec STY-571) that preferentially blocks Abl kinase activity to combat chronic myelogenous leukemia (11).
Modular domains also regulate serine or threonine phosphorylation events. This proceeds through pThr and pSer binding modules such as WW and Forkhead homology–associated (FHA) domains and 14-3-3 proteins (12, 13). The transient nature of these protein-protein interactions implies that these modules must perform a balancing act. Their affinity for a particular binding site must not be too high, or binding will not be regulated by phosphorylation. Yet SH2, WW, or FHA domains must also recognize adjacent residues to allow discrimination between different phosphorylated sites. Furthermore, these modules must display sufficiently high off-rates for rapid and reversible signal transduction. Certain modules exhibit specificities for phosphoinositides phosphorylated at different sites within the inositol ring. A hallmark of these domains is the ability to target their host proteins to specific subcellular localizations, for example, through the recognition of phosphoinositides that mark particular membranes. FYVE domains frequently recognize phosphatidylinositol 3-phosphate [PI(3)P], and thereby direct proteins to endosomes, whereas pleckstrin homology (PH) domains can recognize phosphatidylinositol 4,5-bisphosphate [PI(4,5)P2] or PI(3,4,5)P3, which localize proteins to the plasma membrane, often in response to PI3K signaling (14, 15). A well-studied example is the protein kinase B (PKB)/Akt protein kinase, and its regulator phosphoinositide-dependent protein kinase 1 (PDK1), both of which have PH domains selective for PI(3,4,5)P3 or PtdIns(3,4)P2 and are thus recruited to the plasma membrane and activated in response to PI3K activity (Fig. 1A). Similarly, PLC-δ has a PH domain that selectively binds PI(4,5)P2 with high affinity, which targets the enzyme to regions of membrane enriched in its phospholipid substrate (16, 17). The exquisite selectivity of some PH domains for different phospholipids makes PH domain–containing proteins sensitive to the activities of enzymes that either phosphorylate or dephosphorylate these sites on the inositol ring, such as PI3K or the lipid phosphatase PTEN (phosphatase and tensin homolog deleted from chromosome 10) (18). Accordingly, these enzymes can modulate the localization of downstream signaling proteins that sense distinct phospholipid products. This provides an effective means of assembling or disassembling signaling complexes in different subcellular compartments.
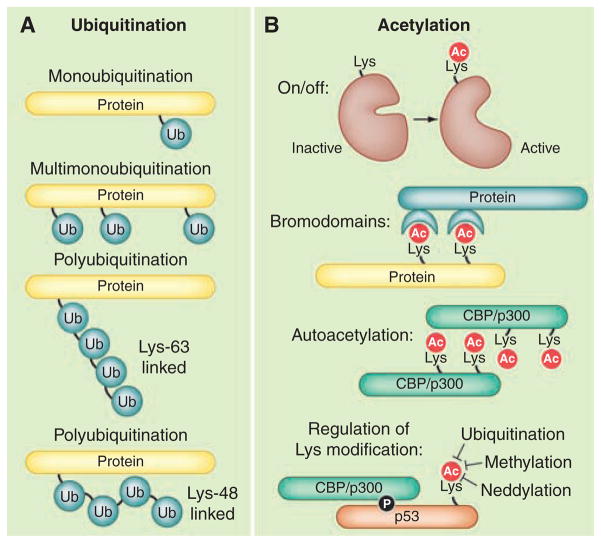
Protein ubiquitination is also used for cell communication (19). This requires the enzymatic attachment of ubiquitin, a 76–amino acid protein tag, to lysine residues on the surface of target substrates (Fig. 2A). Polyubiquitin chains are formed where each ubiquitin molecule is linked through an isopeptide bond to a lysine (K) residue (such as K48, K63 or K11) within another ubiquitin molecule (Fig. 2A). Poly-ubiquitination often leads to the degradation of target proteins by the 26S proteosome. However, the ligation of mono-ubiquitin and di-ubiquitin chains also mediates other cellular functions (20). At least 20 structurally distinct ubiquitin-binding domains (UBDs) are embedded in a variety of proteins, which interpret information conferred by protein ubiquitination in a manner that is reminiscent of phosphotyrosine signaling (21). There are ~600 ubiquitin E3 ligases and more than 90 deubiquitinating proteinases encoded by the human genome. In comparison, there are 523 human protein kinases and 138 protein phosphatases (22). Given such comparable numbers, it is not surprising that ubiquitination and phosphorylation enzymes act synergistically in macromolecular complexes. However, there are added features in events controlled by ubiquitination. For example, UBDs generally bind ubiquitin with micromolar affinities (23). Consequently, these protein-protein interactions are readily reversible and bestow an element of inherent instability within the networks they assemble. This may explain why UBD proteins often control transient cellular processes such as endosomal sorting, vesicular trafficking, and events leading to autophagy (24, 25). It is also worth noting that ubiquitin-like molecules such as SUMO (small ubiquitin-like modifier), Nedd8, and ISG15 are often used as covalent tags to modulate protein function and localization (20, 26).
Fig. 2.
Regulation by ubiquitination or acetylation. These covalent modifications are frequently used for cell communication. (A) Ligation of ubiquitin (Ub) can occur in different patterns on proteins. (B) Acetylation in different contexts serves as an enzymatic on/off switch; for recruitment of bromodomain proteins; (autoacetylation) to induce protein-protein interactions; or for regulation of further lysine modifications.
The ε-amino group of lysine can also undergo reversible acetylation, a versatile form of covalent modification used in different contexts, but often to evoke changes in the activity of histones and transcription factors [Fig. 2B; (27)]. These events are catalyzed by enzymes, often called histone acetyltransferases (HATs), and reversed by histone deaceylases (HDACs). In its simplest form, acetylation functions as an on/off switch to inhibit enzymes such as acetyl-CoA (coenzyme A) synthase and nitric oxide synthase [Fig. 2B; (28)]. Acetylation-dependent protein recruitment of bromodomains, a domain that recognizes acetylated lysine residues, is prevalent in proteins involved in chromatin remodeling (Fig. 2B). In other contexts, autoacetylation initiates dimerization of proteins with intrinsic HAT activity such as the transcriptional co-activator CBP/p300 (Fig. 2B). Finally, lysine acetylation can prevent ubiquitination of the same side-chain, which can be used to prolong the lifetime of proteins that are subject to ubiquitin-proteosome–mediated degradation (Fig. 2B). A classic example is regulation of tumor suppressor protein and transcription factor p53. Under normal conditions, p53 is poly-ubiquitinated and rapidly degraded by the 26S proteosome (29). However, a phosphorylation-acetylation cascade favors the stabilization of p53 in response to DNA damage. When cells are exposed to DNA damage, the phosphorylation of p53 by stress-activated kinases allows its association with p300, which in turn acetylates lysine residues to protect the tumor suppressor from ubiquitin-proteosome–mediated degradation (30).
Thus, distinct covalent modifications can be used in an integrated manner to facilitate the signal-dependent recruitment of proteins. Acetylation of specific lysines can also be used to prevent other major posttranslational modifications. For example, acetylation of individual lysine residues can abolish substrate recognition sites for basophilic protein kinases or, as is the case with p53, occupy ε-amino groups that would otherwise be available for ubiquitination or methylation (Fig. 2B). Thus, the initial pattern and type of posttranslational modifications determine the signaling fate of a given protein, whether it is activation, translocation, or proteosomal destruction. Undoubtedly, high-resolution mass spectrometry will prove to be the best way to explore this phenomenon. A recent study that resolved lysine acetylation profiles of 1750 proteins demonstrated that changes in the amount and frequency of this covalent modification alter a variety of signaling fates, including ubiquitin-mediated degradation and phosphorylation-dependent interactions with 14-3-3 (31).
Processing Signals Through Preassembled Multiprotein Complexes
The passage of signals through preassembled multiprotein signaling complexes is another means of managing intracellular communication. Configuring enzymes in this manner not only enhances the precision of information flow but also improves the fidelity of cell signaling events by clustering successive enzymes into a transduction pathway (Fig. 3A). Commonly, intermediate enzymes in such pathways exhibit restricted substrate specificities and limited spheres of action. In fact, their only true substrate may be the next enzyme in the cascade.
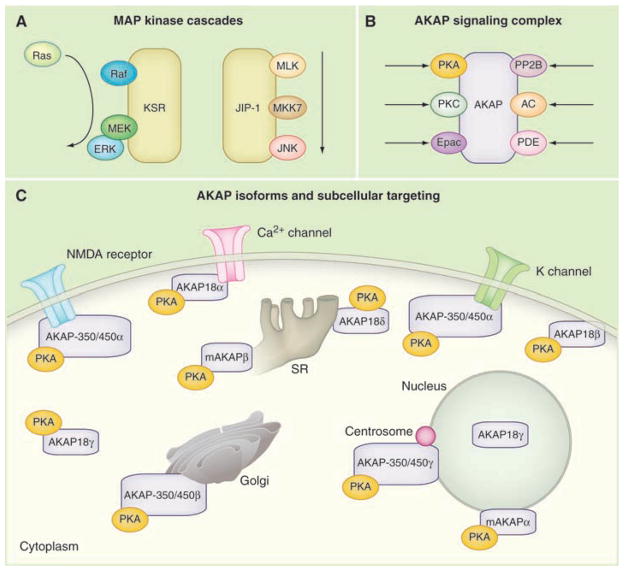
Fig. 3.
Signal transduction through preassembled enzyme complexes. (A) The linear flow of information through the MAP kinase cascades organized by kinase suppressor of Ras (KSR) and JNK interacting protein (JIP). (B) A-kinase anchoring proteins (AKAPs) organize protein kinase A (PKA), guanine nucleotide exchange factors (Epac) and phosphodiesterases (PDE) into cAMP-responsive complexes. Anchoring of certain protein phosphatases (PP2B) and PKC broaden AKAP function. (C) PKA anchoring to various AKAP isoforms targets the kinase to defined subcellular locations. Diagram of a prototypic cell showing the targeting of PKA via AKAP18, AKAP350/450, and mAKAP variants. NMDA, N-methyl-D-aspartate.
This is the case for mitogen-activated protein (MAP) kinases, which form three enzyme-regulatory cascades. Extracellular signals trigger these cascades by stimulating the first member of the pathway, a MAP kinase kinase kinase (MAPKKK). Activated MAPKKKs phosphorylate mitogen-activated protein kinase kinases called MAPKKs or MEKs. This intermediary enzyme phosphorylates the mitogen-activated kinases (MAPKs). The terminal enzyme is then free to act on various downstream targets, including other protein kinases, transcriptional factors, and cytoskeletal components. In mammalian cells, scaffold proteins such as KSR and MP-1 bring together different combinations of MAP kinases to facilitate their activation and sequester these signaling units (Fig. 3A). Likewise, the linear flow of information through the Jun kinase cascade is enhanced by a family of Jun N-terminal kinase (JNK) interacting proteins called JIPs (32). The rationale for such an intricate means of organization lies in how signals are transferred from one enzyme to the next. For example, recruitment of the MAPKKK Raf to the KSR scaffold optimally positions the enzyme in proximity to its target substrate MEK (33, 34). Phospho-MEK is then able to relay the signal to the MAPK (Erk1/2), which involves dual phosphorylation of a threonine and tyrosine in the activation loop of the MAPK. This favors the rapid dissemination of information from one enzyme to the next.
Another useful property of enzyme scaffolding is to segregate enzymes in a manner that prevents indiscriminate cross talk. This is particularly important in unicellular organisms such as yeast where mating, invasive growth, and the response to high osmolarity are regulated by distinct MAPK pathways that share a common MAPKKK called Sterile 11 (Ste11). Segregation of Ste11 activity involves binding to scaffolding proteins such as Pbs2 and Sterile 5 (Ste5). Recruitment of Ste11 into the osmosensing pathway requires interaction with Pbs2 (35). This chimeric protein scaffolds Ste11 and encodes its downstream target, the MAPKK. In contrast, Ste5 organizes Ste11, and the kinases Ste7 and Fus3, to direct signals through the yeast mating pathway (36). Ste5 also facilitates the activation of its kinase-binding partners; a regulatory domain in Ste5 that catalytically unlocks the Fus3 kinase for phosphorylation by Ste7 (37). Thus, Ste5 not only organizes successive components of a yeast MAPK cascade but allosterically modifies the conformation of its bound kinases, making them more amenable to activation (37).
Enzyme Regulation by Subcellular Localization
Compartmentalization of enzymes in proximity to substrates is another means of spatially restricting cell-signaling events. Accordingly, a plethora of kinase- and phosphatase-binding proteins tether their enzyme-binding partners to sites where they can preferentially receive activating signals and be close to selected substrates. Prime examples remain the type 1 phosphatase (PP1) targeting subunits and A-kinase anchoring proteins (AKAPs). The concept of phosphatase targeting was proposed about the same time as the kinase-anchoring hypothesis as a means to generate substrate specificity for second messenger–regulated phosphorylation events (38). The glycogen-particle associated protein GM was the first PP1-targeting subunit to be identified. GM and a functionally related molecule called PTG coordinate signaling complexes that influence glycogen metabolism. Modulation of targeted PP1 activity involves a K/R-V/I-X-F (F, Phe; I, Ile; K, Lys; R, Arg; V, Val; and X, any amino acid) motif in GM that inserts into a groove distal to the active site of the enzyme (39). Peptides encompassing this region are sufficient to displace the PP1 catalytic subunit from GM and abolish the preferential dephosphorylation of glycogen-associated substrates. Thus, GM not only targets PP1 but also allosterically regulates phosphatase activity. The RVxF motif has now been identified in more than 50 potential PP1-targeting subunits. As an example, the muscle-specific phosphatase holoenzyme (PP1-M) contains a targeting subunit called M110/MBS that directs phosphatase activity toward a select group of muscle proteins, including myosin and possibly moesin. Additionally, M110/MBS nucleates a signaling complex with the guanosine 3′,5′-monophosphate (cGMP)–dependent kinase (PKG) and the Rho GTPase. Mobilization of the second messenger cGMP activates PKG that, in turn, phosphorylates M110/MBS to trigger events that lead to smooth-muscle relaxation (40).
Spatial organization of the cAMP-dependent protein kinase (PKA) holoenzyme [consisting of a regulatory (R) subunit dimer and two catalytic (C) subunits] is achieved through interaction with AKAPs (Fig. 3B). High-affinity interaction with PKA is mediated by an amphipathic helix on the AKAP that inserts into a hydrophobic pocket formed by the R-subunit dimer (41, 42). Targeting determinants within the anchoring protein confer the subcellular localization of PKA-AKAP complexes to specific organelles. Mammalian genomes encode about 20 AKAP genes that generate ~75 alternately spliced transcripts. Consequently, multiple variants and differentially targeted isoforms of the same anchoring protein are often expressed within the same cell (Fig. 3C). This increases the repertoire of intracellular PKA anchoring sites and provides a means to restrict action of this broad-spectrum protein kinase toward only a few of its potential substrates. Some AKAPs can also interface with other cAMP signaling elements including adenylyl cyclases, phosphodiesterases, and Epac guanine nucleotide exchange factors (43, 44). Live-cell imaging approaches have shown that these anchored phosphodiesterases participate in negative-feedback loops that locally suppress adenosine 3′,5′-monophosphate (cAMP). Accordingly, these cAMP-responsive units generate local fluctuations in cAMP and concomitant pulses of PKA or Epac activity. Thus, AKAPs appear to orchestrate all aspects of cAMP signaling (Fig. 3B).
A broader role for AKAPs in the spatial organization of cell signaling events became apparent when it was shown that they interact with other regulatory enzymes (Fig. 3B). For example, AKAP79/150 interacts with PKA, the calcium/phospholipid-dependent protein kinase C (PKC) and calmodulin-dependent phosphatase PP2B (45). This implies that second messenger signals that control the phosphorylation and second messenger signals that favor the dephosphorylation of a target substrate pass through the same AKAP complex. This type of regulation may be particularly important for the control of rapid signaling events such as the modulation of neuronal ion channels (Fig. 3C). Strategic use of distinct anchored enzyme combinations provides another way to expand the repertoire of cellular events that the same AKAP modulates. For example, different enzyme combinations anchored to AKAP79/150 modulate the activity of two neuronal ion channels: AMPA (α-amino-3-hydroxy-5-methyl-4-isoxazole propionic acid)–type glutamate receptors and muscarine-sensitive potassium channels (46). In hippocampal neurons, AKAP79/150 coordinates PKA and PP2B modulation of AMPA currents, while any AKAP79/150-associated PKC remains inactive in this process. In contrast, AKAP79/150 enables PKC to facilitate M-current regulation in SCG (superior cervical ganglia) neurons, while PKA and PP2B appear to be non-essential (46).
Unlike scaffolding proteins, which process information in a linear manner (Fig. 3A), the combinatorial assembly of AKAP-enzyme complexes permits the integration and dissemination of multiple signals (Fig. 3B). Although the contextual cues that drive the preferential assembly of distinct AKAP complexes are unclear, one possibility is that the initial binding event of the anchoring protein with its target substrate promotes a succession of conformational changes that select the recruitment of the next binding partners. However, cotranslational assembly of protein complexes through localized protein synthesis, species-specific or cell type–specific expression of particular binding partners may further influence the composition of these “context-dependent” signaling networks.
Temporal Control of Signaling Pathways
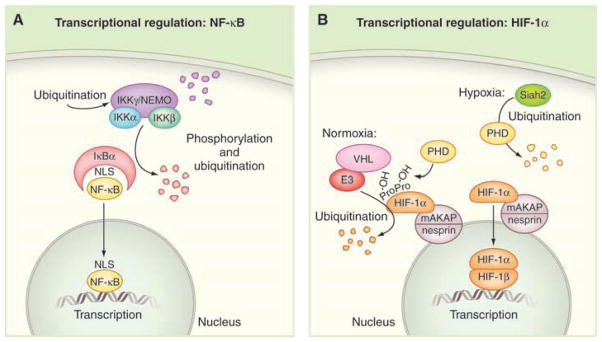
Changes in composition or the amount of enzyme complexes over time also modulate cellular events. This often involves phosphorylation, ubiquitin-mediated degradation, and translocation of signaling components. Nuclear factor κB (NF-κB) is a transcription factor that regulates expression of genes involved in inflammation, apoptosis, and tumorigenesis. Activation of the NF-κB involves the phosphorylation and ubiquitination of several components [Fig. 4A; (47)]. In the absence of stimuli, NF-κB is kept from the nucleus by its interaction with the inhibitory partner IκB. Upon stimulation, IκB is phosphorylated, resulting in subsequent ubiquitination and degradation. The degradation exposes a nuclear localization sequence (NLS) on NF-κB, favoring its translocation to the nucleus. The IκB kinase complex, composed of two catalytic subunits (Iκκα and Iκκβ) and the regulatory subunit (Iκκγ/NEMO), regulates the phosphorylation of IκB (48). Stimulation of the NF-κB pathway results in the ubiquitination of NEMO and activation of the catalytic subunits (Fig. 4A). The enzyme then phosphorylates IκB, resulting in the recruitment of the E3 ubiquitin ligase SCF-βTRCP. This promotes Lys48-linked ubiquitination of IκB, leading to its degradation (49). Because ubiquitination plays a central role in NF-κB activation, its removal by deubiquitinating enzymes is critical to the down-regulation of the NF-κB signal. The cylindromatosis tumor suppressor protein (CYLD) removes Lys63-linked chains from several NF-κB pathway members, including Iκκγ/NEMO, to regulate the duration of NF-κB activation (22). Likewise, dephosphorylation of the IκB kinase complex by protein phosphatase 2A further attenuates the NF-κB response (47). Thus, synchronized ubiquitination and phosphorylation events can exert precise temporal control on a signaling pathway. Yet, the expression of NF-κB target genes often occurs 4 to 6 hours after agonist stimulation. This suggests that a rate-limiting step appears to be the time it takes for NF-κB to translocate from the cytoplasm into the nucleus.
Fig. 4.
Temporal regulation of signaling events. (A) Schematic diagram of the NF-κB pathway; individual protein components are indicated. (B) The concerted actions of oxygen-sensing enzymes and ubiquitin E3 ligases control the stability of the hypoxia-inducible factor protein HIF-1α. Schematic diagram of the HIF-1α pathway under normoxic (left) and hypoxic (right) conditions; individual protein components are indicated.
In oxygen-sensitive tissues, transcriptional responses can occur in minutes rather than hours. The hypoxia-inducible factor 1α (HIF-1α) is rapidly induced in cardiomyocytes and certain tumors in response to reduced intracellular oxygen (50). Under normoxic conditions, the abundance of HIF-1α is kept low through its ubiquitin-mediated proteosomal degradation. This process is initiated by the hydroxylation of two conserved proline residues (Pro402 and Pro564) in HIF-1α, by oxygen-sensitive prolyl hydroxylases (PHDs) [Fig. 4B; (51)]. The hydroxylated proline residues constitute a binding site for the von Hippel–Lindau protein (pVHL), which is part of a complex that ubiquitinates HIF-1α and targets it for degradation by the proteosome (52). During hypoxia, the continual destruction of HIF-1α is halted by the enzymatic activity of PHD that ceases in the absence of oxygen, and a ubiquitin E3 ligase (seven in absentia homolog 2, Siah2) that ubiquitinates PHDs and targets them for proteosomal degradation [Fig. 4B; (53)]. Collectively, these mechanisms terminate the continual destruction of HIF-1α, which allows the protein to form a stable heterodimeric complex with the HIF-1β subunit. The HIF-1α–HIF-1β heterodimer accumulates in the nucleus and initiates transcription of proangiogenic, metabolic, and antiapoptotic genes that promote cell survival. Recent evidence suggests that the anchoring protein (mAKAP) organizes ubiquitin E3 ligases that manage the stability of HIF-1α (54). In cardiomyocytes, depletion of mAKAP or disruption of its targeting to nesprin, a protein that forms the outer ring of the nuclear pore complex, alters the stability of HIF-1α and activation of genes associated with hypoxia. Anchoring of an oxygen-sensitive, ubiquitin-mediated destruction complex at the nuclear pore optimizes temporal control HIF-1α to suppress the hypoxic response. Yet when a hypoxic environment prevails, the transcription factor is released from this anchored complex and immediately translocates to its site of action inside the nucleus (Fig. 4B).
Where Is This Taking Us?
In the past decade, we have witnessed unparalleled advances in our understanding of cell signaling. These include the advent of kinase inhibitor drugs; an appreciation of how phosphorylation, ubiquitination, and acetylation events instigate protein-protein interactions; and the realization that enzyme compartmentalization determines signaling specificity. Technological advances in mass spectrometry, high-throughput genomic sequencing, genetically encoded fluorescent proteins, RNA interference, and live-cell imaging have helped us understand how cellular information is resolved in space and time. If a desirable goal is to deftly manipulate the enzyme activity in space and time, then the future may lie in the burgeoning field of synthetic biology. By taking advantage of the existing knowledge of modular domains, investigators are already generating synthetic molecules that redirect signaling in situ. Prime examples include artificial guanine nucleotide exchange factors designed to “rewire” actin reorganization and alter cell morphology (55), or the design of light-activated Rac1 GTPases that permit laser-induced membrane ruffling at any point in the cell (56). Undoubtedly, these and other innovative approaches will assist our ongoing molecular exploration of the cell.
Acknowledgments
We acknowledge C. Pawson and M. Milnes for evaluation of this manuscript and L. Langeberg for design of the figures. J.D.S. is supported by NIH grant GM48231 and Leducq transatlantic network 06CVD02. T.P. is supported by the National Cancer Institute of Canada, Canadian Cancer Society, Canadian Institutes for Health Research (grants 6849 and 57793), Genome Canada, and a Terry Fox Program Project grant.
References and Notes
- 1.Sadowski I, Stone JC, Pawson T. Mol Cell Biol. 1986;6:4396. doi: 10.1128/mcb.6.12.4396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mayer BJ, Hamaguchi M, Hanafusa H. Cold Spring Harb Symp Quant Biol. 1988;53:907. doi: 10.1101/sqb.1988.053.01.104. [DOI] [PubMed] [Google Scholar]
- 3.Songyang Z, et al. Mol Cell Biol. 1994;14:2777. doi: 10.1128/mcb.14.4.2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kouhara H, et al. Cell. 1997;89:693. doi: 10.1016/s0092-8674(00)80252-4. [DOI] [PubMed] [Google Scholar]
- 5.Ostman A, Heldin CH. Adv Cancer Res. 2007;97:247. doi: 10.1016/S0065-230X(06)97011-0. [DOI] [PubMed] [Google Scholar]
- 6.Fawcett JP, et al. Proc Natl Acad Sci USA. 2007;104:20973. doi: 10.1073/pnas.0710316105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jones N, et al. Nature. 2006;440:818. doi: 10.1038/nature04662. [DOI] [PubMed] [Google Scholar]
- 8.Filippakopoulos P, et al. Cell. 2008;134:793. doi: 10.1016/j.cell.2008.07.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moarefi I, et al. Nature. 1997;385:650. doi: 10.1038/385650a0. [DOI] [PubMed] [Google Scholar]
- 10.Young MA, Gonfloni S, Superti-Furga G, Roux B, Kuriyan J. Cell. 2001;105:115. doi: 10.1016/s0092-8674(01)00301-4. [DOI] [PubMed] [Google Scholar]
- 11.Druker BJ, et al. N Engl J Med. 2001;344:1038. doi: 10.1056/NEJM200104053441402. [DOI] [PubMed] [Google Scholar]
- 12.Yaffe MB, et al. Cell. 1997;91:961. doi: 10.1016/s0092-8674(00)80487-0. [DOI] [PubMed] [Google Scholar]
- 13.Yaffe MB, Cantley LC. Nature. 1999;402:30. doi: 10.1038/46925. [DOI] [PubMed] [Google Scholar]
- 14.Hayakawa A, et al. J Biol Chem. 2004;279:5958. doi: 10.1074/jbc.M310503200. [DOI] [PubMed] [Google Scholar]
- 15.Lemmon MA. Nat Rev Mol Cell Biol. 2008;9:99. doi: 10.1038/nrm2328. [DOI] [PubMed] [Google Scholar]
- 16.Alessi DR, et al. Curr Biol. 1997;7:261. doi: 10.1016/s0960-9822(06)00122-9. [DOI] [PubMed] [Google Scholar]
- 17.Cross DA, Alessi DR, Cohen P, Andjelkovich M, Hemmings BA. Nature. 1995;378:785. doi: 10.1038/378785a0. [DOI] [PubMed] [Google Scholar]
- 18.Lee JO, et al. Cell. 1999;99:323. doi: 10.1016/s0092-8674(00)81663-3. [DOI] [PubMed] [Google Scholar]
- 19.Ciechanover A, Elias S, Heller H, Ferber S, Hershko A. J Biol Chem. 1980;255:7525. [PubMed] [Google Scholar]
- 20.Grabbe C, Dikic I. Science. 2008;322:872. doi: 10.1126/science.1166845. [DOI] [PubMed] [Google Scholar]
- 21.Hicke L, Schubert HL, Hill CP. Nat Rev Mol Cell Biol. 2005;6:610. doi: 10.1038/nrm1701. [DOI] [PubMed] [Google Scholar]
- 22.Sowa ME, Bennett EJ, Gygi SP, Harper JW. Cell. 2009;138:389. doi: 10.1016/j.cell.2009.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Polo S, et al. Nature. 2002;416:451. doi: 10.1038/416451a. [DOI] [PubMed] [Google Scholar]
- 24.Kirkin V, McEwan DG, Novak I, Dikic I. Mol Cell. 2009;34:259. doi: 10.1016/j.molcel.2009.04.026. [DOI] [PubMed] [Google Scholar]
- 25.Raiborg C, Stenmark H. Nature. 2009;458:445. doi: 10.1038/nature07961. [DOI] [PubMed] [Google Scholar]
- 26.Hurley JH, Lee S, Prag G. Biochem J. 2006;399:361. doi: 10.1042/BJ20061138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Basu A, et al. Proc Natl Acad Sci USA. 2009;106:13785. doi: 10.1073/pnas.0906801106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Strahl BD, Allis CD. Nature. 2000;403:41. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 29.Vogelstein B, Lane D, Levine AJ. Nature. 2000;408:307. doi: 10.1038/35042675. [DOI] [PubMed] [Google Scholar]
- 30.Barlev NA, et al. Mol Cell. 2001;8:1243. doi: 10.1016/s1097-2765(01)00414-2. [DOI] [PubMed] [Google Scholar]
- 31.Choudhary C, et al. Science. 2009;325:834. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 32.Whitmarsh AJ, Cavanagh J, Tournier C, Yasuda J, Davis RJ. Science. 1998;281:1671. doi: 10.1126/science.281.5383.1671. [DOI] [PubMed] [Google Scholar]
- 33.Muller J, Ory S, Copeland T, Piwnica-Worms H, Morrison DK. Mol Cell. 2001;8:983. doi: 10.1016/s1097-2765(01)00383-5. [DOI] [PubMed] [Google Scholar]
- 34.Rajakulendran T, Sahmi M, Lefrancois M, Sicheri F, Therrien M. Nature. 2009;461:542. doi: 10.1038/nature08314. [DOI] [PubMed] [Google Scholar]
- 35.Posas F, Saito H. Science. 1997;276:1702. doi: 10.1126/science.276.5319.1702. [DOI] [PubMed] [Google Scholar]
- 36.Choi KY, Satterberg B, Lyons DM, Elion EA. Cell. 1994;78:499. doi: 10.1016/0092-8674(94)90427-8. [DOI] [PubMed] [Google Scholar]
- 37.Good M, Tang G, Singleton J, Remenyi A, Lim WA. Cell. 2009;136:1085. doi: 10.1016/j.cell.2009.01.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cohen P, Cohen TW. J Biol Chem. 1989;264:21435. [PubMed] [Google Scholar]
- 39.Egloff MP, et al. EMBO J. 1997;16:1876. doi: 10.1093/emboj/16.8.1876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Surks HK, et al. Science. 1999;286:1583. doi: 10.1126/science.286.5444.1583. [DOI] [PubMed] [Google Scholar]
- 41.Gold MG, et al. Mol Cell. 2006;24:383. doi: 10.1016/j.molcel.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 42.Kinderman FS, et al. Mol Cell. 2006;24:397. doi: 10.1016/j.molcel.2006.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dodge-Kafka KL, et al. Nature. 2005;437:574. doi: 10.1038/nature03966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bauman AL, et al. Mol Cell. 2006;23:925. doi: 10.1016/j.molcel.2006.07.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Klauck TM, et al. Science. 1996;271:1589. doi: 10.1126/science.271.5255.1589. [DOI] [PubMed] [Google Scholar]
- 46.Hoshi N, Langeberg LK, Scott JD. Nat Cell Biol. 2005;7:1066. doi: 10.1038/ncb1315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Karin M, Cao Y, Greten FR, Li ZW. Nat Rev Cancer. 2002;2:301. doi: 10.1038/nrc780. [DOI] [PubMed] [Google Scholar]
- 48.Mercurio F, et al. Science. 1997;278:860. doi: 10.1126/science.278.5339.860. [DOI] [PubMed] [Google Scholar]
- 49.Rahighi S, et al. Cell. 2009;136:1098. doi: 10.1016/j.cell.2009.03.007. [DOI] [PubMed] [Google Scholar]
- 50.Keith B, Simon MC. Cell. 2007;129:465. doi: 10.1016/j.cell.2007.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Berra E, et al. EMBO J. 2003;22:4082. doi: 10.1093/emboj/cdg392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.George DJ, Kaelin WG., Jr N Engl J Med. 2003;349:419. doi: 10.1056/NEJMp030061. [DOI] [PubMed] [Google Scholar]
- 53.Nakayama K, et al. Cell. 2004;117:941. doi: 10.1016/j.cell.2004.06.001. [DOI] [PubMed] [Google Scholar]
- 54.Wong W, Goehring AS, Kapiloff MS, Langeberg LK, Scott JD. Sci Signal. 2008;1:ra18. doi: 10.1126/scisignal.2000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bashor CJ, Helman NC, Yan S, Lim WA. Science. 2008;319:1539. doi: 10.1126/science.1151153. [DOI] [PubMed] [Google Scholar]
- 56.Wu YI, et al. Nature. 2009;461:104. doi: 10.1038/nature08241. [DOI] [PMC free article] [PubMed] [Google Scholar]