Abstract
Due to the large amount of new papers regularly entering the MEDLINE database, there is an ongoing effort to design tools that help indexing this new material. Here we investigate the hypothesis that past indexing information coming from referencing and authoring links can be used for this purpose. Using a JAMA-based subset of MEDLINE, we designed ranking scores which rely on this information; given a new article, the aim of these scores is to build an ordered list of MeSH terms that should be used to index this article. Evaluation measures on an independent, 1000-document data set are given. Comparison with equivalent works shows benefits in recall, F-measure and mean average precision. Moreover, cited articles and authors’ past articles contribute to seven of the top ten ranking features, supporting our hypothesis. Further improvements and extensions to this work are exposed in the conclusion.
Introduction
MEDLINE indexing involves assigning new articles a set of MeSH descriptors (MeSH terms), possibly along with qualifiers (sub-headings)1. This task is currently manually performed by librarians of the US National Library of Medicine (NLM indexers). Due to the high growth rate of MEDLINE, automatic or semi-automatic tools aiming at assisting indexers are helpful. The NLM indexing initiative’s Medical Text Indexer2 (hereafter MTI) is an indexing tool currently in place at the NLM. It makes Heading and Subheading assignment suggestions using information coming from MetaMap3 tagging of the abstract and title and from the Related Citations2. Other approaches use statistics- and machine-learning-based techniques, yet others use learning-free techniques4.
There are two broad families of techniques. The first family uses only features of the indexed document, whereas the second family uses some kind of structural properties of the database such as neighborhood relationships between documents. The latter approach gives rise to the notion of an Information Hyperspace5. Approaches such as MTI belong to both families. More recent works clearly emphasize the gain of taking into account the relational structure of literature databases that is brought in by co-authoring links, to infer features of documents (nodes of the network) and to suggest new links as well6. Besides, there is a longstanding literature on citation analysis, whose recent developments emphasize the combination of content, author and citation analysis7.
Here we want to take advantage of information preexisting inside MEDLINE, that comes from the established indexing of the previous publications of the authors and of the referenced citations. To do so we have used a Machine Learning technique to create ranking functions which when applied to a new entry propose an ordered list of candidate Main Headings. We evaluate its results using several standard measures.
Material and Methods: Material
Our work is based on the statistical analysis of a set of articles indexed in MEDLINE and for which access to the full text was possible. We have drawn our sample from the Journal of the American Medical Association (JAMA, http://jama.ama-assn.org/), downloading all available on-line material from 1998 to 2008 where a REFERENCES section was available. This choice was motivated by the facts that the format of the articles was HTML (thus sparing us the often tricky work of decoding formats such as PDF), and that the structure of the articles was homogeneous and could be easily processed automatically. Each article is identified by its PubMed ID (PMID).
Methods
We model the indexing task as a supervised learning problem. Its goal is to determine whether the association of a MeSH term to an article is suitable or not. We characterize such an association by a vector of features which takes into account both the contents of the article and the network of co-authorship and citations.
Representation
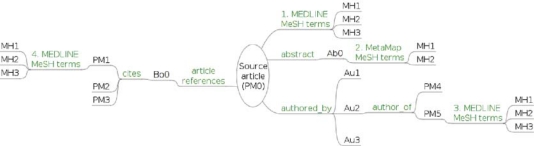
Figure 1 illustrates the local network around a source article in MEDLINE, which we now describe in detail. We applied the following processes to each article in the initial sample from JAMA:
Gather the Main Headings;
Tag the abstract text with MetaMap; we only kept the MeSH terms;
- Parse the REFERENCES section to gather the cited articles; for those indexed by MEDLINE, obtain their PMIDs thanks to straightforward regular expressions (e.g., “link_type=MED” and “access_num=(\d+)”). Then for each cited PMID:
- get its MeSH indexing terms with the Entrez Programming Utilities8;
- for each of these terms compute statistics related to its usage frequency as a descriptor of the references and to which section of the main article (Abstract, Introduction, Methods, Results, Discussion) the references indexed by the current term were used in.
- Parse the list of authors’ names. Then for each author name:
- using Entrez gather the set of MEDLINE indexed articles that were (co-)signed by authors with this name, prior to the date of the source article (only past data must be used in this modeling process);
- then gather the Main Headings of each of these articles;
- compute a series of summary statistics for each of these MeSH terms, essentially related to the past global usage frequencies of the terms by the set of authors (and homonyms) of the main article.
Figure 1:
Different paths for associating MeSH terms to a MEDLINE article. PM=PMID; MH=Main Heading; Au=Author; Ab=Abstract of article; Bo=Body of article
In some cases names are ambiguous in the sense that different authors can share the same name. This typically occurs for example for short last names originating from Asia. We were not able to disambiguate the names here, and we had to stay at the level of homonymous name sets. We believe this limitation has a small negative impact on the efficiency of the new attributes.
In summary for each article of the sample we collected a set of MeSH terms coming from 4 distinct sources (see Figure 1): (1) the MEDLINE Main Headings of the article, (2) the MeSH terms found in the abstract by MetaMap, (3) the MEDLINE Main Headings assigned to previous works by the authors or their homonyms, and (4) the MEDLINE Main Headings of the references cited in the article. Each of these terms was endowed by a set of features including:
the sources where the term was found: Index, MetaMap, Author names, Cited articles. A term is most often found in more than one source, so this information is supported by a set of 4 Boolean variables;
the hierarchical type of the term’s concept from the UMLS Semantic Network (STY)9 and the SNOMED CT axis it belongs to;
some usage frequencies of the term by the authors, and what we call an Inverse Usage Index (IUI) defined as where N is the total number of occurrences of the term in the authors’ work and D is the total number of occurrences of all the terms computed over the whole experimental sample;
using the information gathered in (3.b) above, the frequencies of usage of the term for indexing any referenced article, and the frequencies of the sections of the main article where the referenced articles indexed by this term were cited.
This process produces an experimental data set, each line of which consists of a couple (PMID, MeSH Term), associated with 50 distinct features. We split this data set into two disjoint sets of PMIDs: one was used in the modeling process to create a ranking function, the other for evaluation to obtain unbiased performance measures.
Ranking function
The ranking function was built through supervised classification. The positive examples were the (PMID, MeSH Term) couples for which the MeSH term was indexing the article in MEDLINE. We chose to use a Gradient Boosting algorithm10, as it is a robust, non-parametric (so assumption free) regression method. It iteratively grows a community of simple learners over adaptively re-sampled training sets, the resulting model being an aggregating formula of all simpler models. The original gbm R package11 was used for that.
Results
Our experimental corpus consisted of 3,213 documents. After data preparation we obtained a data set of 2,487,683 (PMID, MeSH Terms) couples (1.71% positive cases). Table 1 shows the distribution of MeSH terms, per source, for articles in our data set. For example the median number of MEDLINE Main Headings of a source article in our corpus is 14. Table 2 displays the results that would be obtained by a classifier returning all collected MeSH terms, evaluating precision and recall on this full list. It is very informative about the contribution of each source. It shows that using only the abstract to choose the indexing terms, whatever the quality of the classifier and the size of the built list, we cannot expect a recall greater than 30% for half the citations. Taking into account also the MeSH Main Headings of the authors’ previous publications and those of the cited articles this median recall could be increased up to 95.24% (even to 100% for more than 25% of the articles), but for a more difficult task as the precision would then drop to less than 2.63% in half the cases.
Table 1:
Distribution of MeSH terms per source. Q1 = first quartile, Q3 = third quartile
| Source | Min. | Q1 | Median | Mean | Q3 | Max. |
|---|---|---|---|---|---|---|
| MEDLINE Indexing Terms of Source Article | 1 | 10 | 14 | 13.28 | 16 | 31 |
| C : Unique Indexing Terms (U.I.T.) of Cited Articles | 2 | 47 | 80 | 86.82 | 114 | 450 |
| A : U.I.T. in the Authors’ previous publications | 2 | 209 | 460 | 808.0 | 990 | 11720 |
| M : MetaMap on abstract | 0 | 27 | 36 | 32.42 | 42 | 76 |
| C ∪ A ∪ M | 3 | 215 | 450 | 773.1 | 940 | 11740 |
Table 2:
Distribution of ‘raw’ recall and precision (with all MeSH terms, unranked), per source
| Source | Precision (%) | Recall (%) | ||||||
|---|---|---|---|---|---|---|---|---|
| Q1 | Median | Mean | Q3 | Q1 | Median | Mean | Q3 | |
| C : Unique Indexing Terms (U.I.T.) of Cited Articles | 9.18 | 13.33 | 15.90 | 19.40 | 66.67 | 80.00 | 75.95 | 90.00 |
| A : U.I.T. in the Authors’ previous publications | 1.12 | 2.31 | 3.90 | 4.66 | 71.43 | 88.24 | 71.35 | 100.00 |
| M : MetaMap on abstract | 8.33 | 12.00 | 12.50 | 16.13 | 17.65 | 30.00 | 29.16 | 41.18 |
| C ∪ A ∪ M | 1.27 | 2.63 | 6.13 | 5.33 | 87.50 | 95.24 | 90.44 | 100.00 |
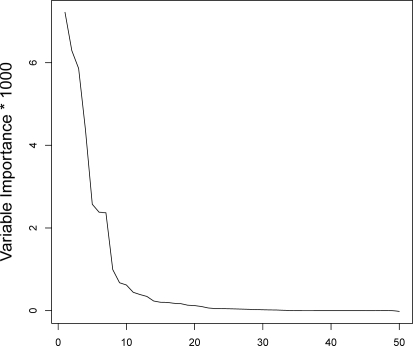
The experimental data set was split into a training data set (2,213 PMID’s) and a validating data set (1,000 PMID’s). Training a Gradient Boosting Machine (1500 decision trees of depth 2; simulated distribution: Bernoulli) over the learning data set with all features as input allowed us to evaluate the importance of each feature in predicting the targeted concept (which is whether or not a MeSH term should be used to index an article, since the ground truth is the existence of the MeSH term as an indexing term for the selected article in MEDLINE. In this process, the actual main headings of the main papers are used only as supervising targets, not as input predictive features.) This is measured as the degradation of the predictive performance when randomly permuting the values of the variable for which importance is computed. Figure 2 advocates using only the ten most influential features (Table 3) out of the 50 initial ones.
Figure 2:
Feature Importance as Predictor
Table 3:
Ten most influential features, ordered by descending importance
| MetaMap_Score: the MetaMap score if the term is found in the abstract, 0 otherwise; |
| FreqDistConcept: the occurrences count of the term divided by the total number of distinct MeSH terms in the cited articles (or 0 when the term does not index any cited article); |
| RefFreq: the proportion of cited articles where the term is used as a Main Heading (or 0 when the term does not index any cited article); |
| RefFreq_Specificity: RefFreq×Specificity; |
| STY: the UMLS Semantic Type of the MeSH term; |
| RefFreq_MeanFreq: RefFreq×MeanFreq; |
| MeanFreq: the past usage frequency of the term by each co-author, averaged over the co-authors set; |
| MetaMap_NbOcc: the number of times the term has been found by MetaMap in the abstract; |
| Specificity: a measure of the specificity of the term for the set of co-authors, that we define as the product of the term’s IUI and the averaged past usage frequency of the term by each co-author; this can be seen as a parallel to the well known tf-idf used in information retrieval12; |
| FreqOccConcepts: the occurrences count of the term divided by the total number of MeSH terms (not necessarily unique) indexing the cited articles (or 0 when the term does not index any cited article). |
We reran the learning algorithm using only these ten features. Comparing the efficiency of the new ranking functions against the ones built with the full feature set showed no performance degradation. The performances of the new ranking functions, expressed as precisions and recalls obtained at increasing ranks of the list of candidate terms built by the ranker for each article, are shown in Tables 4 and 5 (up to rank 25). As these values vary from one document to the other, we give dispersion metrics emphasizing this variability (the quartiles) along with the more classical mean.
Table 4:
Ranking Precisions up to rank 25
| Rank | Min. | Q1 | Median | Mean | Q3 | Max. |
|---|---|---|---|---|---|---|
| 1 | 0.0 | 100.0 | 100.0 | 92.4 | 100.0 | 100.0 |
| 2 | 0.0 | 100.0 | 100.0 | 87.9 | 100.0 | 100.0 |
| 3 | 0.0 | 66.7 | 100.0 | 83.3 | 100.0 | 100.0 |
| 4 | 0.0 | 75.0 | 100.0 | 79.5 | 100.0 | 100.0 |
| 5 | 0.0 | 60.0 | 80.0 | 76.4 | 100.0 | 100.0 |
| 10 | 0.0 | 50.0 | 70.0 | 62.0 | 80.0 | 100.0 |
| 15 | 0.0 | 40.0 | 53.3 | 50.8 | 66.7 | 93.3 |
| 20 | 0.0 | 30.0 | 45.0 | 42.9 | 55.0 | 85.0 |
| 25 | 4.0 | 28.0 | 40.0 | 36.9 | 48.0 | 80.0 |
Table 5:
Ranking Recalls up to rank 25
| Rank | Min. | Q1 | Median | Mean | Q3 | Max. |
|---|---|---|---|---|---|---|
| 1 | 0.0 | 5.6 | 7.1 | 7.6 | 8.3 | 50.0 |
| 2 | 0.0 | 11.1 | 13.3 | 14.5 | 16.7 | 50.0 |
| 3 | 0.0 | 15.0 | 18.8 | 20.4 | 23.1 | 75.0 |
| 4 | 0.0 | 20.0 | 25.0 | 25.6 | 30.8 | 80.0 |
| 5 | 0.0 | 23.5 | 28.6 | 30.4 | 35.7 | 100.0 |
| 10 | 0.0 | 38.5 | 46.7 | 48.0 | 57.1 | 100.0 |
| 15 | 0.0 | 50.0 | 57.1 | 58.2 | 69.2 | 100.0 |
| 20 | 0.0 | 55.6 | 66.7 | 65.2 | 75.3 | 100.0 |
| 25 | 20.0 | 60.0 | 70.6 | 70.0 | 81.2 | 100.0 |
We also provide the 11-point average precision computed on the test data set (Table 6). It appears that the value of the top precision (precision at recall=0) is 0.962. Besides, we also evaluated the Mean Average Precision (MAP) to 0.254.
Table 6:
11-point average precision & MAP
| Recall(%) | 0 | 10 | 20 | 30 | 40 | 50 |
| Prec.(%) | 96.2 | 95.1 | 90.2 | 83.0 | 76.4 | 68.2 |
| Recall(%) | 60 | 70 | 80 | 90 | 100 | |
| Prec.(%) | 58.4 | 47.7 | 36.2 | 23.8 | 13.7 | |
MAP = 25.4 %
Discussion
Tables 4 and 5 are to be interpreted as if the ranking functions were to be used in a recommendation tool during manual indexing. In particular they give hints about the efficiency of the produced lists given their sizes. For example, for lists of 10 items the recall would be greater than 46.7% in half of the cases, and greater than 57.1% in a quarter of the cases. At the same time, the precision would be greater than 70% in half of the cases, and greater than 80% in a quarter of the cases.
Our work follows the same spirit as MTI, but using different information sources (Title, Abstract and Related Citations for MTI, Abstract and past indexing information in our case), and both works target recommendation systems (MTI being nowadays a fully operational tool). So it makes sense to try to compare both approaches as a way to estimate the worthiness of our specific predictive data set. MTI provides both Main Heading and Subheading assignment recommendations whereas we considered the Main Heading assignment task only. Nevertheless, a past evaluation of MTI13 provides measures where only Main Heading assignment is taken into account. At rank 25, the recall value was 0.81 and the precision value was 0.11. In our case, the mean recall at rank 25 is 0.70, but the mean precision is higher at 0.37 (Tables 4 and 5) showing that we favor precision over recall. The corresponding F-measures computed by us for both cases are Fβ =2 = 0.356 for MTI and Fβ =2 = 0.594 for the present work. Nevertheless, let us stress that these numbers must be read with caution, as both experiments may not be fully comparable: the corpora are different (homogeneous in our case), and the mentioned study13 is based on a specifically designed human judgment experiment whereas our experiment follows a machine learning methodology.
In an interesting and quite different approach4 the Main Heading assignment problem is tackled without using any learning technique nor any external information source, so as to avoid the issue of obtaining training data. As it can be viewed as a point of view opposite to ours, a comparison with this work makes sense too. The reported best top precision was 0.914, and the best averaged precision was 0.182, whereas our best top precision is 0.962 and our mean averaged precision is 0.254 (Table 6). Here again these better figures are to be taken cautiously.
Finally, Table 3 shows that each source contributes to the selected features: Authors’ previous work in Ref-Freq_Specificy, RefFreq_MeanFreq, MeanFreq and Specificity; Cited Articles in FreqDistConcept, Ref-Freq, RefFreq_Specificy, RefFreq_MeanFreq and FreqOccConcepts; and Abstract in MetaMap_Score and MetaMap_NbOcc. Combined with the above results, this supports the hypothesis which underlies our approach, namely, that MeSH terms which index cited work and past work by the authors in MEDLINE help to index the present article. Still to be investigated, UMLS Semantic Type contribution may be linked to the fact that score boosting may be beneficial to some specific terms such as those identified as chemical13.
Conclusion
The MEDLINE indexing problem is sometimes viewed as a multilabel assignment one, which makes it hard due to the large number of MeSH terms. Here we adopted a much simpler point of view, where the ranking functions are applied on a term-by-term basis, each term being scored independently of the others.
The aim of this study was purely to estimate the value of new data sources (namely past indexing information) in a MEDLINE indexing process. As it, it did not target the evaluation of different Machine Learning algorithms, and the comparison with previous approaches aimed only at measuring the worthiness of these new predictive features. So far the results seem encouraging, and we are planning some next steps. Among them we want to use a larger and more diverse data set (PubMed Central is an obvious target). We also want to use article titles and information coming from related citations, along with our specific information sources, to obtain a more precise idea of the gain of our specific source of information if added to MTI. Ambiguity in author names is a point we will have to cope with, probably thanks to a prior analysis of the co-authoring network. Eventually, once the worthiness of these new attributes is strongly assessed, we will be able to evaluate different machine Learning algorithms on them.
References
- 1.Medical Subject Headings [WWW page] Bethesda, Maryland; 2009. Accessed March 6, 2010. Available from: http://www.nlm.nih.gov/mesh/meshhome.html.
- 2.Aronson AR, Bodenreider O, Chang F, Humphrey SM, Mork JG, Nelson SJ, et al. Proc AMIA Symp. 2000. The NLM Indexing Initiative; pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 3.Aronson AR. Effective Mapping of Biomedical Text to the UMLS Metathesaurus: The MetaMap Program. Proc AMIA Symp. 2001. pp. 17–21. [PMC free article] [PubMed] [Google Scholar]
- 4.Ruch P, Baud R, Geissbühler A. Learning-Free Text Categorization. In: Dojat M, Keravnou E, Barahona P, editors. Proc. AIME 2003. No. 2780 in LNAI. Springer; 2003. pp. 199–204. [Google Scholar]
- 5.Lin JJ, DiCuccio M, Grigoryan V, Wilbur WJ. Navigating information spaces: A case study of related article search in PubMed. Inform Process Manag. 2008;44(5):1771–1783. [Google Scholar]
- 6.Chang J, Blei DM. Hierarchical Relational Models for Document Networks. Ann Appl Stat. 2010. In press. Available from: http://arxiv.org/abs/0909.4331v2.
- 7.Yang Z, Hong L, Davison BD. Topic-driven Multi-type Citation Network Analysis. In: Grefenstette G, editor. Proc RIAO. Paris: 2010. [Google Scholar]
- 8.Entrez Programming Utilities [WWW page]; 2009. Accessed March 6, 2010. Available from: http://www.ncbi.nlm.nih.gov/entrez/query/static/eutils_help.html.
- 9.Bodenreider O.The Unified Medical Language System (UMLS): integrating biomedical terminology Nucleic Acids Res 2004January1(32(Database issue)):D267–D270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Friedman JH. Greedy Function Approximation: A Gradient Boosting Machine. Ann Stat. 2000;29:1189–1232. [Google Scholar]
- 11.Ridgeway G. gbm: Generalized Boosted Regression Models; 2007. R package version 1.6-3. Available from: http://www.i-pensieri.com/gregr/gbm.shtml.
- 12.Spärck Jones K. A Statistical Interpretation of Term Specificity and its Application in Retrieval. J Doc. 1972;28(1):11–21. [Google Scholar]
- 13.Aronson AR, Mork JG, Gay CW, Humphrey SM, Rogers WJ. The NLM Indexing Initiative’s Medical Text Indexer. Studies in Health Technology and Informatics. 2004. pp. 268–272. [PubMed]