Abstract
Measurement of 8-hydroxy-2′-deoxyguanosine (8-OH-dGuo) in DNA by high-performance liquid chromatography/mass spectrometry (LC/MS) was studied. A methodology was developed for separation by LC of 8-OH-dGuo from intact and modified nucleosides in DNA hydrolyzed by a combination of four enzymes: DNase I, phosphodiesterases I and II and alkaline phosphatase. The atmospheric pressure ionization-electrospray process was used for mass spectral measurements. A stable isotope-labeled analog of 8-OH-dGuo was used as an internal standard for quantification by isotope-dilution MS (IDMS). Results showed that LC/IDMS with selected ion-monitoring (SIM) is well suited for identification and quantification of 8-OH-dGuo in DNA at background levels and in damaged DNA. The sensitivity level of LC/IDMS-SIM was found to be comparable to that reported previously using LC-tandem MS (LC/MS/MS). It was found that approximately five lesions per 106 DNA bases can be detected using amounts of DNA as low as 2 µg. The results also suggest that this lesion may be quantified in DNA at levels of one lesion per 106 DNA bases, or even lower, when more DNA is used. Up to 50 µg of DNA per injection were used without adversely affecting the measurements. Gas chromatography/isotope-dilution MS with selected-ion monitoring (GC/IDMS-SIM) was also used to measure this compound in DNA following its removal from DNA by acidic hydrolysis or by hydrolysis with Escherichia coli Fpg protein. The background levels obtained by LC/IDMS-SIM and GC/IDMS-SIM were almost identical. Calf thymus DNA and DNA isolated from cultured HeLa cells were used for this purpose. This indicates that these two techniques can provide similar results in terms of the measurement of 8-OH-dGuo in DNA. In addition, DNA in buffered aqueous solution was damaged by ionizing radiation at different radiation doses and analyzed by LC/IDMS-SIM and GC/IDMS-SIM. Again, similar results were obtained by the two techniques. The sensitivity of GC/MS-SIM for 7,8-dihydro-8-oxoguanine was also examined and found to be much greater than that of LC/MS-SIM and the reported sensitivity of LC/MS/MS for 8-OH-dGuo. Taken together, the results unequivocally show that LC/IDMS-SIM is well suited for sensitive and accurate measurement of 8-OH-dGuo in DNA and that both LC/IDMS-SIM and GC/IDMS-SIM can provide similar results.
INTRODUCTION
Oxygen-derived free radicals such as the hydroxyl (OH) radical generate multiple modifications in DNA including base and sugar lesions, strand breaks and DNA–protein crosslinks (1–3). This type of DNA damage, which is also called oxidative DNA damage or free radical-induced DNA damage, is implicated in mutagenesis, carcinogenesis and aging (4). Accurate measurement of DNA lesions is required for understanding the mechanisms of oxidative DNA damage and its cellular repair, and the role of this damage in certain diseases. DNA damage can be measured by a variety of techniques including high-performance liquid chromatography (LC) with electrochemical detection (ECD) (reviewed in 5–7), LC-tandem mass spectrometry (LC/MS/MS) (8–10) and gas chromatography-MS (GC/MS) (reviewed in 6,11). LC-ECD and LC/MS/MS were mainly used only for the measurement of 8-hydroxy-2′-deoxyguanosine (8-OH-dGuo) (5–10). On the other hand, GC/MS is capable of measuring multiple modified bases from all four DNA bases in a single DNA sample (6,11). Of the multiple products generated by free radicals in DNA, 7,8-dihydro-8-oxoguanine (8-hydroxyguanine; 8-OH-Gua, also called 8-oxoGua) was extensively studied as a biomarker, due to its mutagenic potential and the facile measurement of its nucleoside 8-OH-dGuo by LC-ECD (5–7). However, the measurement of a single product as a biomarker may be misleading because of the free radical-induced generation of multiple products from all four bases in DNA at the same time (1–3). The advantage of measuring multiple modified bases in DNA was recently demonstrated (12,13).
There is a significant discrepancy between endogenous levels of 8-OH-Gua or 8-OH-dGuo in DNA reported in the literature (5,7). This suggests variability in measurements depending on the laboratory and/or the measurement technique. Recently, a European Standards Committee for Oxidative DNA Damage (ESCODD) was established to resolve problems associated with the measurement of 8-OH-Gua or 8-OH-dGuo. Two trials were undertaken in which samples of 8-OH-dGuo and calf thymus DNA from a stock source were measured in different laboratories using a variety of analytical techniques. Published results showed significant differences between the levels of 8-OH-Gua or 8-OH-dGuo reported from various laboratories (14,15).
Due to its biological importance and broad use as a biomarker, it is important to measure 8-OH-Gua or 8-OH-dGuo accurately and reliably. In a comparative study, we recently reported the measurement of 8-OH-Gua by GC/MS following hydrolysis either by Escherichia coli Fpg protein or by formic acid. These two procedures of hydrolysis yielded similar levels of 8-OH-Gua in DNA, suggesting that no artifactual formation of 8-OH-Gua from free guanine occurred during derivatization of formic acid-hydrolysates (16). An artifactual formation of 8-OH-Gua during derivatization of formic acid-hydrolysates prior to GC/MS-analysis was alleged (reviewed in 17). However, recent studies showed that artifactual formation is dependent on the laboratory and procedures used, and can be prevented (14,15,18–21).
In the present study, we applied LC/MS and GC/MS to measure 8-OH-dGuo and 8-OH-Gua, respectively, in DNA with the aim to compare these two techniques. Previously, LC/MS/MS was used for the measurement of 8-OH-dGuo following enzymatic hydrolysis of DNA to nucleosides and in urine (8–10). A further aim of the present study was to compare the results obtained with LC/MS to those previously published using LC/MS/MS. We report on the measurement by LC/MS and GC/MS of background levels of 8-OH-dGuo in commercial calf thymus DNA and in DNA of cultured human cells and its levels in calf thymus DNA γ-irradiated in aqueous solution.
MATERIALS AND METHODS
Materials
Materials for GC/MS were obtained as described (22). Calf thymus DNA and phosphodiesterase I were obtained from Sigma Chemical Company (St Louis, MO). DNase I, phosphodiesterase II and alkaline phosphatase were purchased from Roche Diagnostics Corporation (Indianapolis, IN). Acetonitrile (HPLC-grade) was from Allied Signal, Inc. Burdick and Jackson (Muskegon, MI). 2′-Deoxyguanosine-1,3,7,9-15N4-(2-amino-15N) (dGuo-15N5) was purchased from Cambridge Isotope Laboratories (Andover, MA). 8-OH-dGuo, 8-hydroxy-2′-deoxyguanosine-8-18O (8-OH-dGuo-18O) and 8-hydroxyguanine-1,3-15N2-(2-amino-15N)-2-13C (8-OH-Gua-15N3-13C) were synthesized by Dr Victor Nelson from Program Resources, Inc. (Frederick, MD) (23). dGuo-15N5, 8-OH-dGuo and 8-OH-dGuo-18O were dissolved in water. 8-OH-Gua-15N3-13C was dissolved in 0.01 N NaOH. The concentration of these compounds was calculated by weight and by measuring their absorption spectra using the following absorption coefficients: dGuo-15N5, 13 000 M–1cm–1 at 254 nm (24); 8-OH-dGuo and 8-OH-dGuo-18O, 9908 M–1cm–1 at 245 nm (25); 8-OH-Gua-15N3-13C, 11 600 M–1cm–1 at 283 nm (18). Water (HPLC-grade) for LC/MS analyses was purchased from J. T. Baker (Phillipsburg, NJ). Water purified through a Milli-Q system (Millipore Corporation, Bedford, MA) was used for all other applications.
Preparation of DNA samples
Calf thymus DNA was dissolved in 10 mM Na-phosphate buffer (pH 7.4; 0.3 mg/ml) at 4°C. The concentration of DNA was determined by UV spectroscopy (1 absorbance unit = 50 µg of DNA/ml). Aliquots of this solution were bubbled with N2O and irradiated with γ-rays in a 60Co γ-source at doses of 5, 10, 20, 40 and 80 Gy (dose rate 35 Gy/min). Irradiated and unirradiated DNA samples were dialyzed against 10 mM Na-phosphate buffer (pH 7.4) for 18 h at 4°C. The buffer solution outside the dialysis bags was changed three times during the course of the dialysis. Aliquots of these solutions containing 100 µg of DNA were dried under vacuum in a SpeedVac.
Cell culture and DNA isolation
HeLa (adenocarcinoma; cervix) cells (ATCC, Manassas, VA) were grown in 175 cm2 flasks in antibiotic-free Dulbecco’s modified Eagle’s medium (Life Technologies, Rockville, MD) containing 10% (v/v) fetal bovine serum (Life Technologies). Media was aspirated off and cells were rinsed with 20 ml of 1× phosphate-buffered saline. Cells were detached by trypsin (Life Technologies) and DNA was isolated using a blood and cell culture DNA maxi kit (Qiagen, Valencia, CA). DNA was recovered by spooling, washed once in 70% ethanol, and then air-dried. After dissolving DNA in 10 mM phosphate buffer (pH 7.4), DNA concentration was determined by UV spectroscopy. DNA concentration was adjusted to 0.3 mg/ml.
Enzymatic hydrolysis of DNA to nucleosides
Hydrolysis of DNA was accomplished by the combined use of four enzymes as described previously (26). An aliquot of 8-OH-dGuo-18O and 100 µl of 2 mM CaCl2 were added to dried unirradiated and irradiated DNA samples (100 µg). The total phosphate buffer concentration was 35 mM (pH 7.4). DNase I (4 U), phosphodiesterase I (0.0032 U), phosphodiesterase II (0.08 U) and alkaline phosphatase (34 U) were added and the samples incubated at 37°C for 6 h. Subsequently, 250 µl of cold ethanol (at –20°C) were added and the samples were kept at –20°C for 30 min. Samples were centrifuged at 15 000 g for 30 min. Supernatant fractions were separated and dried under vacuum in a SpeedVac. Samples were dissolved in 100 µl of water, vortexed vigorously for 5 min and then centrifuged at 15 000 g for 30 min. Approximately 60 µl of the clear solution were separated and used for injection onto the column of the LC/MS instrument.
Hydrolysis of DNA with formic acid
Aliquots of 8-OH-Gua-15N3-13C (2 pmol) and dGuo-15N5 (2.5 µmol) were added to unirradiated and irradiated DNA samples (100 µg) as internal standards and the samples were dried under vacuum in a SpeedVac. The use of dGuo-15N5 permits determination of the DNA amount by GC/isotope-dilution MS (GC/IDMS). Upon hydrolysis, this compound yields guanine-15N5, which is used as an internal standard for quantification of guanine in DNA, and consequently for the determination of the DNA amount (21). The results of DNA determination by this method and by UV absorbance measurements correlated well with each other. DNA samples were hydrolyzed and derivatized for GC/MS analysis as described (16).
Hydrolysis of DNA with E.coli Fpg protein
8-OH-Gua was excised from DNA by hydrolysis with E.coli Fpg protein as described (16). An aliquot of 8-OH-Gua-15N3-13C (2 pmol) was added to the hydrolysates. DNA was precipitated with ethanol and the samples centrifuged at 15 000 g. Supernatant fractions were separated, lyophilized and derivatized as described (16).
Analysis by LC/MS
DNA samples hydrolyzed to nucleosides were analyzed by LC/MS with selected-ion monitoring (SIM) using a 1100 Series liquid chromatograph-mass selective detector (Agilent Technologies, Rockville, MD) equipped with a UV-spectrophotometer and an automatic injector. The atmospheric pressure ionization-electrospray (API-ES) process in the positive ionization mode was used. The flow and temperature of the drying gas (nitrogen) were 10 l/min and 350°C, respectively. The nebulizing gas pressure was 172 kPa. The capillary potential was 4000 V. The fragmentor potential was varied from 60 to 120 V to find the optimal conditions. Electron multiplier potential was 2600 V. Quadrupole temperature was 99°C. An Ultra IBD C18-reversed-phase column (25 cm × 2.1 mm i.d., 5 µm particle size) (Restek Corporation, Bellefonte, PA) was used. A guard column packed with the same stationary phase (1 cm × 2.1 mm i.d.) was attached to the column head. The solvents A and B were water and acetonitrile, respectively. A gradient of 0.5% of solvent B per minute was used. The flow rate was 0.2 ml/min. The column temperature was kept at 30°C. An aliquot of 2–10 µl of hydrolyzed DNA samples (1 µg of DNA/µl) was injected onto the column for each analysis.
Analysis by GC/MS
The derivatized samples of formic acid- and Fpg protein-hydrolysates were analyzed by GC/MS-SIM as described previously (16,21).
RESULTS
The objective of this study was to develop a methodology using LC/MS to measure 8-OH-dGuo in DNA and establish whether this technique is suitable for this purpose in terms of sensitivity and accuracy. A further objective was to compare the results obtained by LC/MS with those obtained by the well-established GC/MS technique. Such a comparative study was essential to establish whether or not both techniques would yield similar results. Thus, an artifactual formation of 8-OH-Gua during derivatization of formic acid-hydrolysates of DNA was recently alleged (17). However, recent studies established that artifactual formation is dependent upon the laboratory and experimental procedures used and can be prevented (14–16,19–21). In the LC/MS procedure, there is no derivatization step and an artifactual formation of 8-OH-dGuo should be limited to DNA isolation (7) and enzymatic hydrolysis that hydrolyzes DNA to nucleosides. It was established that formic acid hydrolysis used for GC/MS analysis does not generate any artifacts (16,27). Using DNA samples from the same source and assuming that enzymatic hydrolysis does not generate artifacts, a comparison of the results obtained by LC/MS and GC/MS should reveal whether the derivatization step prior to GC/MS analysis generates any artifacts. A recent study using E.coli Fpg protein or formic acid to excise 8-OH-Gua from DNA prior to GC/MS analysis suggested that, if performed under proper experimental conditions, derivatization does not generate artifacts (16). Furthermore, we wanted to compare LC/MS with LC/MS/MS, which was previously used for the measurement of 8-OH-dGuo in DNA (8,9), in terms of sensitivity and capability to measure this lesion in DNA at low levels.
Measurement of 8-OH-dGuo in DNA by LC/MS
Chromatographic conditions for LC were established by using several types of reversed-phase columns and solvent conditions. The best separation of 8-OH-dGuo from intact nucleosides was obtained using a 25 cm × 2.1 mm (5 µm particle size) column. The elution order of intact nucleosides was typical of reversed-phase columns (28) with 2′-deoxycytidine eluting first (7.2 min) followed by 2′-deoxyguanosine (13.1 min), 2′-deoxythymidine (14.4 min) and 2′-deoxyadenosine (19.5 min). 2′-Deoxyinosine eluting at 12.2 min was also observed in enzymatic hydrolysates of DNA, probably due to the deamination of 2′-deoxyadenosine by deaminases present in alkaline phosphatase (29). 8-OH-dGuo eluted at 17.4 min, in between 2′-deoxythymidine and 2′-deoxyadenosine and was completely separated from them. The complete separation of 8-OH-dGuo from 2′-deoxyguanosine with a difference of 4 min between elution times was important because of the reported possible oxidation of 2′-deoxyguanosine to 8-OH-dGuo during the mass spectrometric process (9,10). Water and acetonitrile were used as solvents. Ammonium acetate buffer (10 mM) was also examined as a solvent instead of water. However, a significant reduction in sensitivity in the response of the mass spectrometer was observed when compared to water. Therefore, water and acetonitrile were used as solvents for all applications. The peak shapes obtained with this solvent system were sharp and symmetrical. This elution system is simple and very well suited for LC/MS.
The API-ES process was used for mass spectral measurements. The positive-ion mass spectrum of 8-OH-dGuo was recorded in the total-ion monitoring mode using a series of fragmentor potentials to establish its fragmentation pattern. The mass spectrum consisted of the protonated molecular ion (MH+) at m/z 284, the protonated free base ion (BH2+) at m/z 168, the sodium adduct ion (MNa+) at m/z 306 and a sugar ion (S+) at m/z 117. The ion intensities depended upon the fragmentor potential. At a fragmentor potential of 100 V, the ion at m/z 168 had the highest abundance (100%) followed by those at m/z 284 (8%), m/z 306 (5%) and m/z 117 (3%). The mass spectrum (not shown) was similar to the positive-ion mass spectrum of 8-OH-dGuo published previously using LC/MS with API-ES (30) and to those obtained using LC/MS/MS (8,9). 8-OH-dGuo-18O, which was used as an internal standard, yielded a similar mass spectrum with MH+ at m/z 286, BH2+ at m/z 170, MNa+ at m/z 308 and S+ at m/z 117.
The measurement of the level of 8-OH-dGuo in enzymatic hydrolysates of DNA samples was performed using the SIM mode. The m/z 168 and 284 ions for 8-OH-dGuo and m/z 170 and 286 ions for its stable isotope-labeled analog 8-OH-dGuo-18O as an internal standard were recorded simultaneously within the appropriate time period, where 8-OH-dGuo eluted. As expected, no difference between the retention times of these two compounds was observed. The fragmentor potential was set to 100 V and this provided the highest sensitivity for the ions at m/z 168 and 170, which were used for quantification of 8-OH-dGuo. First, a calibration plot was obtained for the response of the mass spectrometer to known quantities of both 8-OH-dGuo and 8-OH-dGuo-18O. Mixtures of these compounds were prepared with the ratio of the molar amount of 8-OH-dGuo to that of 8-OH-dGuo-18O varying between 0.357 and 11.25. These mixtures were analyzed by LC/MS-SIM and the ions at m/z 168 and 170 were recorded. The ratios of ion currents were plotted as a function of the ratios of molar amounts. Figure 1 illustrates the calibration plot. A linear relationship was obtained with a slope of 1.04. For actual measurements of 8-OH-dGuo in hydrolysates of DNA, the amount of the internal standard was chosen according to the limits of the ratio of molar amounts shown in Figure 1 in order to ascertain the linear response of the mass spectrometer.
Figure 1.
The calibration plot for the ratio of the ion-currents at m/z 168 (8-OH-dGuo) and m/z 170 (8-OH-dGuo-18O) versus the ratio of the molar amounts of 8-OH-dGuo and 8-OH-dGuo-18O. All values represent the average (± standard deviation) of three independent measurements. For these measurements, the injected amounts of 8-OH-dGuo and 8-OH-dGuo-18O varied from 0.1 and 0.28 nmol (ratio = 0.357), respectively, to 0.45 and 0.04 nmol (ratio = 11.25), respectively.
Sensitivity of LC/MS
Figure 2 illustrates ion-current profiles at m/z 168 and 170 recorded during the LC/MS-SIM analysis of an enzymatic hydrolysate of 10 µg of calf thymus DNA. The profile of the ion at m/z 168 corresponds to ~400 fmol of 8-OH-dGuo on the column. The sensitivity of the instrument was tested under the set of conditions used in this study by analyzing 1–5 µg of DNA. Figure 3 illustrates the ion-current profile at m/z 168 obtained by injecting 2 µg of DNA onto the LC column and represents ~70 fmol of 8-OH-dGuo on the column. The ratio of signal to background was ~5 (Fig. 3). This sensitivity level is ~3.5-fold lower than that reported for 8-OH-dGuo as measured by LC/MS/MS (20 fmol) (9). The same paper reported a sensitivity level of 5 pmol (5000 fmol) for 8-OH-dGuo when LC/MS-SIM mode of measurement was used. The sensitivity level obtained in the present study using LC/MS-SIM is ~70-fold greater than this. When 1 µg of DNA was injected, a signal-to-noise ratio of ~2.5 was found, corresponding to ~35 fmol of 8-OH-dGuo (data not shown). This was the limit of detection. However, the peak shape was not appropriate for an accurate integration. The ion-current profile at m/z 168 shown in Figure 3 represents approximately 10 lesions per 106 DNA bases (0.0325 nmol/mg of DNA). This is the level of this compound detected in calf thymus DNA. This and other levels of 8-OH-dGuo (or 8-OH-Gua) obtained under various conditions are given below. It should be pointed out that the level shown in Figure 3 was obtained with 2 µg of DNA, which contained 10 lesions/106 DNA bases. This suggests that, if DNA contained 8-OH-dGuo at a level of, for example, one to two lesions per 106 DNA, it would be possible to quantify 8-OH-dGuo by analyzing ≥10 µg of DNA by LC/IDMS-SIM. In the present study, up to 50 µg of DNA were injected on the LC column and no abnormalities in the analysis of 8-OH-dGuo were observed.
Figure 2.
Ion-current profiles at m/z 168 (8-OH-dGuo) and m/z 170 (8-OH-dGuo-18O) recorded during LC/IDMS-SIM analysis of calf thymus DNA (10 Gy) hydrolyzed by a combination of four enzymes. An aliquot of 10 µg of hydrolyzed DNA was injected on the LC column.
Figure 3.
The ion-current profile at m/z 168, which corresponds to ~70 fmol of 8-OH-dGuo. The amount of DNA injected onto the LC column was 2 µg.
Sensitivity of GC/MS
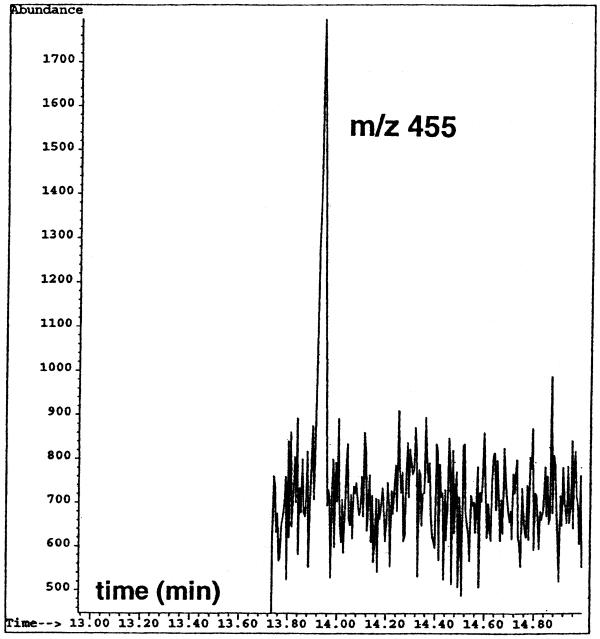
Figure 4 illustrates the profile of the ion at m/z 455, which is the molecular ion of the trimethylsilyl derivative of 8-OH-Gua (31,32). This was recorded during the GC/MS-SIM analysis of a formic acid-hydrolysate of calf thymus DNA. The amount of DNA on the column for this analysis was ~0.1 µg, when a split ratio of 1:75 at the injection port of the GC-instrument was used during the injection of the sample onto the GC column. The ion-current profile in Figure 4 corresponds to ~3 fmol of the trimethylsilyl derivatives of 8-OH-Gua on the GC column and represents approximately 10 lesions per 106 DNA bases (0.0325 nmol/mg of DNA). This sensitivity level is much greater than those obtained by LC/MS-SIM (this study) and LC/MS/MS (9), respectively. Furthermore, it should be pointed out that much less DNA amount was used to obtain this level of sensitivity than the amounts of DNA used for LC/MS-SIM or LC/MS/MS measurements.
Figure 4.
The ion-current profile at m/z 455 recorded during the GC/IDMS-SIM analysis of a derivatized formic acid-hydrolysate of DNA. The signal corresponds to ~3 fmol of 8-OH-Gua. The amount of DNA on the GC-column was ~0.1 µg for this measurement after splitting the sample with a split ratio of 1:75. The original amount of the sample injected onto the column was 4 µl containing ~8 µg of DNA.
Levels of 8-OH-dGuo and 8-OH-Gua in DNA as measured by LC/MS and GC/MS
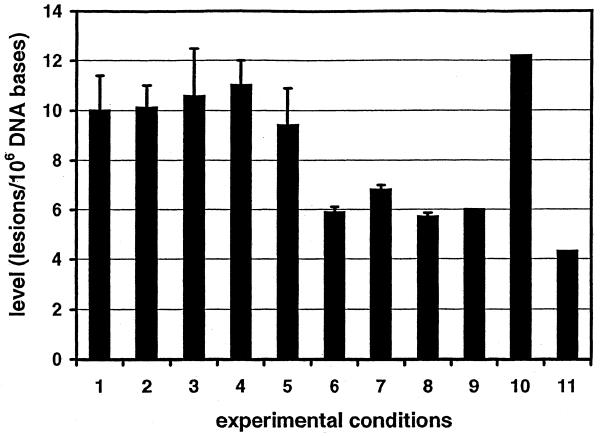
Aliquots of DNA samples were analyzed by LC/IDMS-SIM following enzymatic hydrolysis and by GC/IDMS-SIM following formic acid or Fpg protein hydrolysis and derivatization. The levels of 8-OH-dGuo or 8-OH-Gua were measured in calf thymus DNA, in DNA isolated from cultured HeLa cells and in calf thymus DNA γ-irradiated in aqueous solution. The background levels obtained under a variety of experimental conditions are shown in Figure 5. Some relevant results previously obtained using LC/MS/MS are also shown for comparison. The level of 8-OH-dGuo measured by LC/IDMS-SIM using enzymatic hydrolysis and that of 8-OH-Gua measured by GC/IDMS-SIM using hydrolysis by formic acid or hydrolysis by Fpg protein were similar, with no statistical difference (Fig. 5, columns 1–3, respectively). These were also similar to the levels that were published from this laboratory using GC/IDMS-SIM with hydrolysis by formic acid or hydrolysis by Fpg protein (columns 4 and 5, respectively). DNA isolated from HeLa cells had significantly lower levels of 8-OH-dGuo (columns 6–8) than those in calf thymus DNA (columns 1–5). As in the case of calf thymus DNA, there was no statistical difference between the level of 8-OH-dGuo in DNA of cultured HeLa cells measured by LC/IDMS-SIM using enzymatic hydrolysis and that of 8-OH-Gua measured by GC/IDMS-SIM using hydrolysis by formic acid or hydrolysis by Fpg protein (columns 6–8). Three levels of 8-OH-dGuo previously measured by LC/MS/MS in calf thymus DNA (9,15) and in rat liver (8) are also shown in Figure 5 for comparison (columns 9–11, respectively). In these cases, no standard deviation or error was given. Therefore, it is not possible to compare these values (columns 9–11) with those from this study, or results published previously (columns 1–8), in terms of statistical difference. In general, these values are similar to those found in this study.
Figure 5.
The level of 8-OH-dGuo measured by LC/IDMS-SIM and the level of 8-OH-Gua measured by GC/IDMS-SIM in DNA under various conditions. Columns 1–3: in calf thymus DNA, measured by LC/IDMS-SIM with hydrolysis by four enzymes, GC/IDMS-SIM with hydrolysis by formic acid and by GC/IDMS-SIM with hydrolysis by Fpg protein, respectively (this work). Columns 4 and 5: in calf thymus DNA, previously measured by GC/IDMS-SIM with hydrolysis by formic acid and by GC/IDMS-SIM with hydrolysis by Fpg protein, respectively (16). Columns 6–8: in DNA isolated from cultured HeLa cells, measured by LC/IDMS-SIM with hydrolysis by four enzymes, GC/IDMS-SIM with hydrolysis by formic acid and by GC/IDMS-SIM with hydrolysis by Fpg protein, respectively (this work). All values in columns 1–8 represent the average (± standard deviation) of three to six independent measurements. Columns 9 and 10: in calf thymus DNA measured by LC/MS/MS (9,15). Column 11: in rat liver measured by LC/MS/MS (8). No standard deviation or standard error was given for the values in columns 9–11.
Samples of calf thymus DNA γ-irradiated in buffered aqueous solution were also analyzed by LC/IDMS-SIM and GC/IDMS-SIM to test whether or not both techniques yield similar results when damaged DNA is used. A dose range from 5 to 80 Gy was used to irradiate DNA solutions. Figure 6 illustrates the dose–yield plots of 8-OH-dGuo and 8-OH-Gua obtained by LC/IDMS-SIM and GC/IDMS-SIM, respectively. An excellent correlation was observed between the two techniques.
Figure 6.
Radiation dose–yield plots of 8-OH-dGuo and 8-OH-Gua in DNA exposed to various doses of ionizing radiation. Circle, 8-OH-dGuo measured by LC/IDMS-SIM with hydrolysis by four enzymes. Square, 8-OH-Gua measured by GC/IDMS-SIM with hydrolysis by formic acid. All values represent the average (± standard deviation) of three independent measurements.
DISCUSSION
The results show that LC/MS-SIM is well suited for the sensitive and accurate measurement of 8-OH-dGuo in DNA. The sensitivity level of this technique measured in this study is ~4-fold lower than the reported sensitivity level of LC/MS/MS and much greater than the level previously reported for LC/MS-SIM (9). It was possible to determine 8-OH-dGuo at a level of six to 10 lesions per 106 DNA bases using 2 µg of DNA. The results suggest that it would be possible to determine this compound at a level of one lesion per 106 DNA bases by using more DNA for analysis. Chromatographic conditions permitted a complete separation of 8-OH-dGuo from all intact nucleosides, especially from 2′-deoxyguanosine. This facilitated a significant reduction in the background noise, increasing the level of sensitivity. The cost of LC/MS equipment is much less than that of LC/MS/MS equipment. This could make this technique more attractive to many laboratories than LC/MS/MS for measurement of 8-OH-dGuo or any other lesions in DNA. The sensitivity level of GC/MS-SIM was determined to be greater than that of LC/MS-SIM and LC/MS/MS. The GC/MS equipment used in this laboratory was >10 years old. State-of-the-art equipment may even provide a greater sensitivity level.
The levels of 8-OH-dGuo and 8-OH-Gua in calf thymus DNA or HeLa DNA measured by LC/IDMS-SIM and GC/IDMS-SIM were similar. This clearly indicates that these two techniques can provide similar results. This fact was unequivocally confirmed by the use of DNA samples γ-irradiated at different radiation doses. An excellent correlation between the results obtained by these two techniques was observed. These results also demonstrate that there was no artifactual formation of 8-OH-Gua during derivatization of formic acid-hydrolysates of DNA for GC/MS analysis under the experimental conditions of this study. This was confirmed by using Fpg protein for excision of 8-OH-Gua from DNA instead of formic acid. The level of 8-OH-Gua obtained by GC/IDMS-SIM using Fpg protein for hydrolysis was similar to levels obtained by LC/IDMS-SIM or GC/IDMS-SIM following hydrolysis by formic acid. Unlike formic acid, this enzyme does not excise the intact bases. Therefore, there is no possibility of artifactual formation of 8-OH-Gua from guanine, which is not present in Fpg protein-hydrolysates of DNA. These results also confirm those recently reported from this laboratory (16). Furthermore, the results show that lyophilization of formic acid-hydrolysates does not generate any artifactual formation of 8-OH-Gua from guanine in contrast to the reported generation of 8-OH-dGuo during lyophilization of DNA solutions (33,34).
It should be pointed out that DNA isolation might also produce artifacts causing formation of 8-OH-dGuo or any other products in DNA (reviewed in 5,7). Prevention of oxidation of DNA by various procedures during DNA isolation was described recently (see for example 5,7,33–36). These procedures included the use of antioxidants and free radical scavengers during DNA isolation and the chaotropic NaI method. In the present study, a commercial kit was used to isolate DNA from HeLa cells according to the procedure recommended by the manufacturer. At present, it is not known whether such a commercial kit would cause artifacts during DNA isolation. However, the level of 8-OH-Gua or 8-OH-dGuo measured in this study by GC/MS or LC/MS in DNA of cultured HeLa cells was similar to most of the previously reported levels of this compound in DNA from other tissues. This indicates that isolation of DNA in this study might have caused a minimum amount of oxidation or no oxidation of guanine in DNA. On the other hand, the objective of the present study was to develop methodologies to measure 8-OH-dGuo in DNA by LC/MS, and not to study the effect of DNA isolation.
Another aspect of the results of this study is the observation that enzymatic hydrolysis of DNA with the use of a combination of four enzymes releases the same level of 8-OH-dGuo from DNA as the level of 8-OH-Gua released by formic acid. The use of even extensively irradiated DNA samples with a high level of damage (650 lesions per 106 DNA bases at 80 Gy) yielded similar results. This clearly indicates that the release of 8-OH-dGuo from DNA by enzymatic hydrolysis was complete under the conditions used in this study, assuming that formic acid hydrolysis completely excises 8-OH-Gua from DNA. Previously, it was unequivocally shown (27,37) that 8-OH-Gua is completely released from DNA by formic acid under the hydrolysis conditions similar to those used in the present study. Our results also show that the use of a combination of four enzymes, DNase I, phosphodiesterases I and II and alkaline phosphatase, for the release of 8-OH-dGuo from DNA is as effective as the commonly-used combination of two enzymes, nuclease P1 and alkaline phosphatase, for this purpose (27,34). This is in agreement with a recent study that showed that the four-enzyme combination was as effective as the two-enzyme combination for complete hydrolysis of 8-OH-dGuo from DNA (34). On the other hand, this is in contrast to a recent claim, which did not provide data on DNA, that the four-enzyme combination might not be suitable for hydrolyzing DNA to nucleosides (38).
Conclusions
The following conclusions can be drawn from the present study.
(i) LC/IDMS-SIM is a well-suited technique for the measurement of 8-OH-dGuo in DNA. The sensitivity of this technique is comparable to that of the LC/MS/MS reported previously.
(ii) Low levels of 8-OH-dGuo in DNA, such as five lesions per 106 DNA bases, can be detected using amounts of DNA as low as 2 µg. Furthermore, the results suggest that this lesion may be quantified in DNA at levels of one lesion per 106 DNA bases, or even lower, when more DNA is used.
(iii) The levels of 8-OH-dGuo measured by LC/IDMS-SIM and 8-OH-Gua measured by GC/IDMS-SIM in DNA at background levels or in damaged DNA were almost identical, indicating that similar results can be obtained by both techniques under the conditions used in the present study.
(iv) Similar results obtained by LC/IDMS-SIM and GC/IDMS-SIM unequivocally show that: (a) enzymatic hydrolysis as used in this study completely excises 8-OH-dGuo from DNA, even from heavily damaged DNA; (b) derivatization of formic acid-hydrolysates does not artifactually generate any 8-OH-Gua from guanine under the conditions of this study. The latter fact is in accord with recently published results from this laboratory.
(v) The sensitivity of GC/IDMS-SIM for measurement of 8-OH-Gua is much greater than that of LC/IDMS-SIM and the reported sensitivity of LC/MS/MS for measurement of 8-OH-dGuo.
Further studies are required to show whether LC/IDMS-SIM is also well suited for sensitive and accurate measurement of other DNA lesions.
Acknowledgments
ACKNOWLEDGEMENTS
We are grateful to Dr Timothy R. O’Connor of City of Hope National Medical Center (Duarte, CA) for the gift of E.coli Fpg protein. Certain commercial equipment or materials are identified in this paper in order to specify the experimental procedures. Such identification does not imply recommendation or endorsement by the National Institute of Standards and Technology, nor does it imply that the materials or equipment identified are necessarily the best available for the purpose.
References
- 1.Téoule R. (1987) Radiation-induced DNA damage and its repair. Int. J. Radiat. Biol., 51, 573–589. [DOI] [PubMed] [Google Scholar]
- 2.Dizdaroglu M. (1992) Oxidative damage to DNA in mammalian chromatin. Mutat. Res., 275, 331–342. [DOI] [PubMed] [Google Scholar]
- 3.Breen A.P. and Murphy,J.A. (1995) Reactions of oxyl radicals with DNA. Free Radic. Biol. Med., 18, 1033–1077. [DOI] [PubMed] [Google Scholar]
- 4.Halliwell B. and Gutteridge,J.M.C. (1999) Free Radicals in Biology and Medicine. Oxford University Press, New York, NY, pp. 351–429.
- 5.Collins A., Cadet,J., Epe,B. and Gedik,C. (1997) Problems in the measurement of 8-oxoguanine in human DNA. Carcinogenesis, 18, 1833–1836. [DOI] [PubMed] [Google Scholar]
- 6.Dizdaroglu M. (1998) Mechanisms of free radical damage to DNA. In Aruoma,O.I. and Halliwell,B. (eds), DNA and Free Radicals: Techniques, Mechanisms and Applications. OIC International, Saint Lucia, pp. 1–24.
- 7.Helbock H.J., Beckman,K.B., Shigenaga,M.K., Walter,P.B., Woodall,A.A., Yeo,H.C. and Ames,B.N. (1998) DNA oxidation matters: The HPLC-electrochemical detection assay of 8-oxo-deoxyguanosine and 8-oxo-guanine. Proc. Natl Acad. Sci. USA, 95, 288–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Serrano J., Palmeira,C.M., Wallace,K.B. and Kuehl,D.W. (1996) Determination of 8-hydroxydeoxyguanosine in biological tissue by liquid chromatography/electrospray ionization-mass spectrometry/mass spectrometry. Rapid Commun. Mass Spectrom., 10, 1789–1791. [DOI] [PubMed] [Google Scholar]
- 9.Ravanat J.-L., Duretz,B., Guiller,A., Douki,T. and Cadet,J. (1998) Isotope dilution liquid chromatography-electrospray tandem mass spectrometry assay for the measurement of 8-oxo-7,8-dihydro- 2′-deoxyguanosine in biological samples. J. Chromatogr., 715, 349–356. [DOI] [PubMed] [Google Scholar]
- 10.Renner T., Fechner,T. and Scherer,G. (2000) Fast quantification of the urinary marker of oxidative stress 8-hydroxy-2′-deoxyguanosine using solid-phase extraction and high-performance liquid chromatography with triple-stage quadrupole mass detection. J. Chromatogr., 738, 311–317. [DOI] [PubMed] [Google Scholar]
- 11.Dizdaroglu M. (1991) Chemical determination of free radical-induced damage to DNA. Free Radic. Biol. Med., 10, 225–242. [DOI] [PubMed] [Google Scholar]
- 12.Podmore I.D., Griffiths,H.R., Herbert,K.E., Mistry,N., Mistry,P. and Lunec,J. (1998) Vitamin C exhibits both a pro-oxidant and antioxidant behavior in vivo. Nature, 392, 559. [DOI] [PubMed] [Google Scholar]
- 13.Rehman A., Collis,C.S., Yand,M., Kelly,M., Diplock,A.T., Halliwell,B. and Rice-Evans,C. (1998) The effect of iron and vitamin C co-supplementation on oxidative damage to DNA in healthy volunteers. Biochem. Biophys. Res. Commun., 246, 293–298. [DOI] [PubMed] [Google Scholar]
- 14.Lunec J. (1998) ESCODD: European Standards Committee on Oxidative DNA Damage. Free Radic. Res., 29, 601–608. [DOI] [PubMed] [Google Scholar]
- 15.ESCODD (2000) Comparison of different methods of measuring 8-oxoguanine as a marker of oxidative DNA damage. Free Radic. Res., 32, 333–341. [DOI] [PubMed] [Google Scholar]
- 16.Rodriguez H., Jurado,J., Laval,J. and Dizdaroglu,M. (2000) Comparison of the levels of 8-hydroxyguanine as measured by gas chromatography mass spectrometry following hydrolysis of DNA by Escherichia coli Fpg protein or formic acid. Nucleic Acids Res., 28, e75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cadet J., Douki,T. and Ravanat,J.-L. (1997) Artifacts associated with the measurement of oxidized DNA bases. Environ. Health Perspect., 105, 1034–1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hamberg M. and Zhang,L.-Y. (1995) Quantitative determination of 8-hydroxyguanine and guanine by isotope dilution mass spectrometry. Anal. Biochem., 229, 336–344. [DOI] [PubMed] [Google Scholar]
- 19.Jenner A., England,T.G., Aruoma,O.I. and Halliwell,B. (1998) Measurement of oxidative DNA damage by gas chromatography-mass spectrometry: Ethanethiol prevents artifactual generation of oxidized DNA bases. Biochem. J., 331, 365–369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.England T.G., Jenner,A., Aruoma,O.I. and Halliwell,B. (1998) Determination of oxidative DNA base damage by gas chromatography-mass spectrometry. Effect of derivatization conditions on artifactual formation of certain base oxidation products. Free Radic. Res., 29, 321–330. [DOI] [PubMed] [Google Scholar]
- 21.Sentürker S. and Dizdaroglu,M. (1999) The effect of experimental conditions on the levels of oxidatively modified bases in DNA as measured by gas chromatography-mass spectrometry: How many modified bases are involved? Prepurification or not? Free Radic. Biol. Med., 27, 370–380. [DOI] [PubMed] [Google Scholar]
- 22.Dizdaroglu M. (1994) Chemical determination of oxidative DNA damage by gas chromatography-mass spectrometry. Methods Enzymol., 234, 3–16. [DOI] [PubMed] [Google Scholar]
- 23.Nelson V.C. (1996) Synthesis of isotopically labelled DNA degradation products for use in mass spectrometric studies of cellular DNA damage. J. Label. Comp. Radiopharm., 38, 713–723. [Google Scholar]
- 24.Fasman G.D. (1983) Handbook of Biochemistry and Molecular Biology, 3rd Edn. CRC Press, Boca Raton, FL, pp. 65–215.
- 25.Cavalieri L.F. and Bendich,A. (1950) The ultraviolet absorption spectra of pyrimidines and purines. J. Am. Chem. Soc., 72, 2587–2594. [DOI] [PubMed] [Google Scholar]
- 26.Dizdaroglu M. (1985) Formation of an 8-hydroxyguanine moiety in deoxyribonucleic acid on γ-irradiation in aqueous solution. Biochemistry, 24, 4476–4481. [DOI] [PubMed] [Google Scholar]
- 27.Ravanat J.-L., Gremaud,E., Markovic,J. and Turesky,R.J. (1998) Detection of 8-oxoguanine in cellular DNA using 2,6-diamino-8-oxopurine as an internal standard for high-performance liquid chromatography with electrochemical detection. Anal. Biochem., 260, 30–37. [DOI] [PubMed] [Google Scholar]
- 28.Pomerantz S.C. and McCloskey,J.A. (1990) Analysis of RNA hydrolyzates by liquid chromatography-mass spectrometry. Methods Enzymol., 193, 796–824. [DOI] [PubMed] [Google Scholar]
- 29.Crain P.F. (1990) Preparation and enzymatic hydrolysis of DNA and RNA for mass spectrometry. Methods Enzymol., 193, 782–790. [DOI] [PubMed] [Google Scholar]
- 30.Reddy D.M. and Iden,C.R. (1993) Analysis of modified deoxynucleosides by electrospray ionization mass spectrometry. Nucl. Nucl., 12, 815–826. [Google Scholar]
- 31.Dizdaroglu M. (1985) Application of capillary gas chromatography-mass spectrometry to chemical characterization of radiation-induced base damage in DNA: implications for assessing DNA repair processes. Anal. Biochem., 144, 593–603. [DOI] [PubMed] [Google Scholar]
- 32.Dizdaroglu M. (1993) Quantitative determination of oxidative base damage in DNA by stable isotope-dilution mass spectrometry. FEBS Lett., 315, 1–6. [DOI] [PubMed] [Google Scholar]
- 33.Gedik C.M., Wood,S.G. and Collins,A.R. (2000) Measuring oxidative damage to DNA; HPLC and the comet assay compared. Free Radic. Res., 29, 605–615. [DOI] [PubMed] [Google Scholar]
- 34.Wood S.G., Gedik,C.M. and Collins,A.R. (2000) Controlled oxidation of calf thymus DNA to produce standard samples for 8-oxo-deoxyguanosine analysis: Effects of freeze-drying, storage and hydrolysis conditions. Free Radic. Res., 32, 327–332. [DOI] [PubMed] [Google Scholar]
- 35.Egil K. and Tyrell,R.M. (1997) Artificial background and induced levels of oxidative DNA base damage in DNA from human cells. Carcinogenesis, 18, 2281–2283. [DOI] [PubMed] [Google Scholar]
- 36.Hofer T. and Möller,L. (1998) Reduction of oxidation during the preparation of DNA and analysis of 8-hydroxy-2′-deoxyguanosine. Chem. Res. Toxicol., 11, 882–887. [DOI] [PubMed] [Google Scholar]
- 37.Nackerdien Z., Olinski,R. and Dizdaroglu,M. (1992) DNA base damage in chromatin of γ-irradiated cultured human cells. Free Radic. Res. Commun., 16, 259–273. [DOI] [PubMed] [Google Scholar]
- 38.Romieu A., Gasparutto,D. and Cadet,J. (1999) Synthesis and characterization of oligonucleotides containing 5′,8-cyclopurine 2′-deoxyribonucleosides: (5′R)-5′,8-cyclo-2′- deoxyadenosine, (5′S)-5′,8-cyclo-2′-deoxyguanosine and (5′R)-5′,8-cyclo-2′-deoxyguanosine. Chem. Res. Toxicol., 12, 412–421. [DOI] [PubMed] [Google Scholar]