Abstract
Natural language processing of clinical notes is challenging due to a high degree of semantic ambiguity. Previous research has uncovered ways to improve disambiguation accuracy using manually created rules of semantic sentence structure. However, applying a natural language processing system in a new clinical domain using this method is very labor intensive. This paper presents an automatic method of developing such disambiguation rules for a wide range of clinical domains. Our rules are based on the co-occurrence patterns of semantic types of terms unambiguously mapped to UMLS concepts by MetaMap. These patterns are combined into a sublanguage semantic schema that can be used by an existing natural language processing system such as MetaMap. The differences of co-occurrence patterns across clinical notes of different domains are presented here as evidence of clinical sublanguages.
Introduction
There are two main approaches to building a natural language processing (NLP) system. The first approach is symbolic, based on manually created language models consisting of syntactic and/or semantic rules. The other is statistical, using machine learning principles to derive a language model from a large set of manually annotated text. Both of these approaches require a high level of human effort. Applying an existing NLP system to a new domain usually leads to a significant decrease in system performance because of the difference in vocabulary and language usage across domains due to sublanguage variations.
Once build, domain adaptation of NLP systems also has been recognized as an expensive and labor-demanding task because it often involves manual annotation of text from the new domain or development of a language model by human experts. Minimizing the level of human involvement in domain adaptation would significantly increase domain portability of an NLP system.
Clinical Sublanguage
The notion of a sublanguage was formally explored by Zellig Harris.1 According to his language theory, the inequalities of the likelihood of word co-occurrence within a specific sentence based on word semantic types limit the possible combinations of words in a sentence. Some word combinations have zero probability and are said to be outside of the syntax of the particular language. In addition to general language constraints, a sublanguage is characterized by a set of constraints that do not hold in general language.
Sublanguages vary in their domain-specific vocabularies, word semantic types, and sentence semantic structure.1 Evidence from limited previous research suggests that the narratives authored by clinicians of different specialties exhibit sublanguage characteristics.2 Once the sublanguage constraints are defined, a more accurate word sense disambiguation can be achieved through the semantic type co-occurrence rules.3,4
Word Sense Disambiguation
Word sense disambiguation (WSD) is a process of selecting an appropriate meaning for a term, in context, from a range of possible meanings for that word. WSD starts with determining a set of possible meanings for each investigated word using an existing dictionary. The Unified Medical Language System (UMLS) Knowledge Sources are often used in the biomedical domain. The UMLS Semantic Network is based on 135 semantic types. Each term in the UMLS Metathe-saurus is assigned one or more semantic types depending on the term meaning, usage, and domain.
Using the semantic type of the context concepts to guide word sense selection has been shown to be an effective way to improve disambiguation accuracy.5 Fan and Friedman exploited unambiguous concepts resulting from MetaMap text processing to create semantic type classifiers for ambiguous terms.6 Several excellent reviews of work in the area of WSD have appeared recently.7,8
Sublanguage-based processing
One of the earliest descriptions of medical language as a sublanguage of general English was presented by Pratt and Pacak in 1969.9 The Linguistic String Project (LSP) at New York University, which drew its theoretical basis from the work of Harris, was originally developed for general English. Later it was expanded by a panel of experts into the medical domain by creating a set of medical concepts, semantic categories, and sentence structures to represent clinical data.10
MedLEE, a highly successful clinical NLP system developed by Carol Friedman,4 traces its roots to the LSP. MedLEE specifies a relatively small number of semantic types that are used to constrain the interpretation of text. Even though the system architecture is quite flexible, making it useful to a broader clinical audience requires lexicon expansion as well as manual creation of disambiguation rules for each clinical domain.
MetaMap
MetaMap is a powerful concept recognition system developed by a team led by Aronson at the National Library of Medicine.11 Its primary aim is to map terms found in abstracts of MEDLINE citations, as well as user queries to concepts in the UMLS Metathesaurus. A recent comprehensive overview of MetaMap system is presented elsewhere.11
Its comprehensiveness, robustness, free availability, and regular updates with the latest version of the UMLS make MetaMap very attractive for potential NLP users. However, in spite of the good coverage of the clinical domain by the UMLS Metathesaurus, MetaMap has not been applied widely to clinical text processing beyond a few research projects. The main deterrent to broad application of MetaMap on clinical narratives is its failure to perform accurate word sense disambiguation. When a term from free text matches multiple UMLS concepts, MetaMap returns a list of all mappings, making information extraction difficult. If MetaMap’s WSD algorithm is used on clinical narratives, it often selects the wrong concept because it was trained on biomedical text.
Providing a simple and effective method to adapt MetaMap WSD to clinical narratives would open the door to a wider MetaMap acceptance for research and clinical system development. To this end we propose an automated method to develop sublanguage a semantic schema (SSS) that, following Harris, consists of a set of domain specific semantic types and word sense disambiguation rules based on semantic type co-occurrence patterns.
Methods
The complete set of all clinical narrative types at our medical center (a large tertiary care teaching hospital) in use during the period January 2007-December 2008 was analyzed by a clinical expert to determine a study subset that was diverse across domains. Note types that consisted mostly of templated information, scanned hand-written documentation, or non-clinical documents were excluded. As a result, a set of 17 representative note types was selected for this study. These note types represented a cross-section of clinical narratives created by clinical personnel that varied by clinical role (physicians, nurses), specialty (Cardiology, Dermatology, Ob/Gyn, Oncology, etc.) and clinical environment (ED, inpatient, outpatient).
A set of 683,125 notes was extracted from the University Hospital Electronic Data Warehouse. Files that were less than 100 bytes in length were excluded because they did not contain clinically relevant information. The remaining 559,029 files were processed by the MetaMap binary (v.2009)12 running locally on a secure, HIPAA-compliant, high-performance compute cluster. MetaMap failed to process some files completely, and we address this in the Limitations section. Those incomplete files were excluded in order to decrease the bias due to the language differences within the notes. The remaining 231,303 files were analyzed for the current project. In addition to the clinical narratives, a random set of 35,000 MEDLINE abstracts published between 2000–2008 was selected and processed by MetaMap. Since the target corpus for MetaMap is the MEDLINE abstracts, we included the abstracts in the analysis to illustrate language differences. To ensure a valid comparison to the clinical texts, abstracts less than 100 bytes and those that failed to be processed completely were excluded.
The MetaMap output was delivered in XML format. It was parsed and all unambiguously mapped terms for each note type were extracted. Previous research with biomedical texts has used a simple definition of unambiguous mappings: those terms mapped to a single concept.6 This definition is not appropriate for clinical data because of their high level of ambiguity, which results in extremely limited mappings. After reviewing several hundreds of clinical text mappings produced by MetaMap, additional rules of unambiguity were derived. We rely on the evaluation metric generated by MetaMap to measure the quality of the match between the term in the analyzed phrase and a Metathesaurus concept.13 In our approach, a mapping is called unambiguous if any of the following conditions are met:
MetaMap produced only one concept match for the term even if variant generation was required to find this match (variants reduce the MetaMap mapping score, but here the mapping is still unambiguous).
MetaMap produced a single identical match except for spelling variation, capitalization, NOS suffixes and inversions such as Cancer, Lung vs. Lung Cancer.
MetaMap produced a single match for the term such that the match evaluation score is either over 900 or, if no mappings over 900 are found, over 800.
For clarity, the following definitions will be used in the remainder of this text:
token - the smallest lexical unit analyzed by MetaMap, such as words, numbers, or punctuation.
term - one or more semantically linked tokens that MetaMap analyzes as a syntactic unit.
mapping - a term that was unambiguously mapped to a UMLS concept using the rules above. A mapping has a UMLS concept identifier and a semantic type associated with it.
ST - the semantic type of the UMLS concept associated with the mapping.
Results
The distribution of several features in the successfully processed files is outlined in Table 1. Initial ST frequency counts revealed that almost all note types had an average of between two and three mappings per sentence, compared to five for MEDLINE abstracts. This observation led us to conclude that evaluating patterns of more than 3 mappings would fail to produce useful patterns, due to sparsity. So for each mapping, three terms before and after a mapping of interest within the sentence were extracted. Relative frequencies of observed sequences of mappings that fell within the evaluation window were calculated. Ambiguously mapped terms were counted as unmapped. The evaluated co-occurrence patterns represented a sequence of mappings and other terms found within the text of each clinical note type. The three most common pattern formats were:
Format 1 - a format where a mapping alternated with another term in a sequence.
Format 2 - a format where one mapping is followed by another term followed by two adjacent mappings.
Format 3 - a format where two adjacent mappings are followed by another term followed by one more mapping.
Table 1.
Pertinent counts for each note type used in this study.
| # | Note Type with associated abbreviation | Note count | Mappings per note | Sentences per note | Mappings per sentence |
|---|---|---|---|---|---|
| 1 | AHP - Admission History and Physical | 16, 846 | 225.2 | 88.2 | 2.6 |
| 2 | ANN - Ambulatory Nursing Note | 28, 515 | 18.1 | 8.5 | 2.1 |
| 3 | BCN - Burn Clinic Note | 5, 237 | 63.9 | 25.3 | 2.5 |
| 4 | CCN - Cardiology Clinic Note | 8, 567 | 137.5 | 69.4 | 2.0 |
| 5 | CMD - Case Management Discharge Plan | 11, 532 | 30.9 | 10.2 | 3.0 |
| 6 | DCN - Dermatology Clinic Note | 2, 262 | 52.4 | 25.2 | 2.1 |
| 7 | DS - Discharge Summary | 25, 853 | 183.4 | 66.5 | 2.8 |
| 8 | EDR - Emergency Dept Report | 681 | 126.6 | 60.0 | 2.1 |
| 9 | FPC - Family Practice Clinic Note | 4, 140 | 72.9 | 33.7 | 2.2 |
| 10 | HOC - Hematology Oncology Clinic | 14, 354 | 148.9 | 60.1 | 2.4 |
| 11 | NCN - Neurology Clinic Note | 8, 547 | 175.7 | 67.4 | 2.6 |
| 12 | OGC - Ob-Gyn Clinic Note | 3, 241 | 106.1 | 42.3 | 2.5 |
| 13 | OR - Operative Report | 32, 475 | 130.0 | 48.4 | 2.7 |
| 14 | OCN - Orthopaedic Clinic Note | 58, 807 | 70.2 | 30.2 | 2.3 |
| 15 | PSC - Plastic Surgery Clinic Note | 1, 512 | 57.4 | 24.5 | 2.3 |
| 16 | RCN - Rheumatology Clinic Note | 7, 439 | 90.4 | 30.8 | 2.9 |
| 17 | SSN - Social Service Note | 1, 295 | 69.4 | 22.5 | 3.1 |
| 18 | MLN - MEDLINE abstracts | 21, 972 | 46.3 | 9.2 | 5.0 |
Table 2 describes these formats and gives examples of patterns and sentence sections that match these formats. Format 1 accounted for the most pattern instances in the analyzed corpus.
Table 2.
Most common pattern formats. The semantic type (ST) element in the table refers to the 4-character abbreviation provided by MetaMap. For the definitions, see the SRDEF file located at http://semanticnetwork.nlm.nih.gov/Download/
|
Format 1 Example: [ST ] |
mapping patient [podg] |
term was |
mapping examined [fndg] |
term and |
mapping treated by [ftcn] |
|
Format 2 Example: [ST ] |
mapping clear [qlco] |
term to |
mapping auscultation [diap] |
mapping bilaterally [spco] |
term in all fields |
|
Format 3 Example: [ST ] |
mapping # grams [qnco] |
mapping daily [tmco] |
term to |
mapping see [acty] |
term if |
Table 3 shows the most common pattern of Format 1 with examples for each note type. Several clinical notes share some of the most common semantic type co-occurrence patterns, which indicates that the sublanguages of those clinical notes are related. On the other hand, some patterns, such as a list of Hazardous or Poisonous Substances (ST: [hops]) in ED Reports, appear frequently in one note type but almost never in other note types. MEDLINE abstracts show a relatively high frequency of patterns that consist of the same semantic type indicating a list of similar concepts, such as chemicals [orch], geographic locations [geoa], proteins [aapp], antibiotics [antb], and genes [gngm] in the following format: mapping, mapping, and/or mapping. Out of these patterns, only a list of Organic Chemicals (orch) appear frequently in clinical notes. In those cases, the Organic Chemical refers to a name of a generic medication.
Table 3.
Examples of the most frequent patterns for each note type.
| Note Type | Pattern Example with [ST] | Relative Frequency |
|---|---|---|
| ANN | tubes labeled [qlco] and sent [acty] to the lab [mnob] | 28.3% |
| BCN | patient [podg] was examined [[fndg] and treated [[ftcn] | 25.9% |
| DCN | arms [blor], hands [bloc] or legs [blor] | 16.2% |
| PSC | pneumonia [dsyn], heart failure [dsyn], stroke [dysn] | 7.9% |
| HOC | lungs [bpoc] are clear [qlco] to auscultation [diap] | 6.4% |
| FPC | lungs [bpoc] are clear [qlco] to auscultation [diap] | 6.2% |
| OGC | extremity [bpoc] tender [qlco] to palpation [diap] | 5.9% |
| CCN | lungs [bpoc] are clear [qlco] to auscultation [diap] | 5.5% |
| CMD | reason [idcn] for admission [hlca] was surgical procedure [topp] | 5.3% |
| OCN | zocor [orch], Norvasc [orch], and diovan [orch] | 4.8% |
| RCN | shoulder [blor], wrist [blor], elbows [blor] | 4.5% |
| NCN | Paxil [orch], Ambien [orch], and nabumetone [orch] | 4.2% |
| AHP | lungs [bpoc] are clear [qlco] to auscultation [diap] | 3.9% |
| EDR | cocaine [hops], methamphetamines [hops] or other [fndg] | 3.8% |
| SSN | continue [idcn] to follow [inpr] the patient [podg] | 3.1% |
| DS | aspirin [orch], phenytoin [orch] and Cozaar [orch] | 3.1% |
| OR | #-year-old [tmco] gravida # [fndg], para # [fndg] | 1.0% |
| MLN | for sophoridine[orch], sophocarpine [orch], and matrine [orch] | 0.4% |
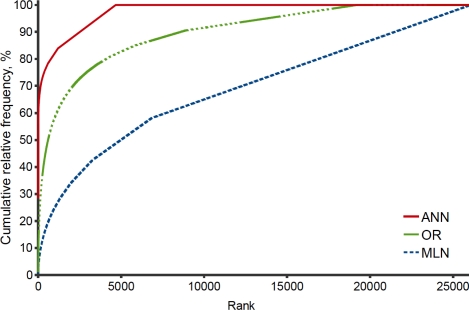
Each pattern that appears frequently in a corpus can be viewed as a semantic co-occurrence rule. A set of rules that describe a large proportion of instances of semantic type sequences in a corpus defines a sublanguage semantic structure. A rule set “growth curve” indicates how many rules describe a given proportion of the patterns found in a corpus sorted by their relative frequency. Figure 1 illustrates the differences in the rule set growth curves for two clinical note types and MEDLINE abstracts. The curves for the other evaluated clinical notes fell between the lines for Ambulatory Nursing Notes (ANN) and Operating Reports (OR) and are not displayed here for readability.
Figure 1.
Format 1 rule set growth curve: clinical notes versus MEDLINE abstracts.
The slope of the distribution of the relative frequency of patterns in MEDLINE is very flat. The most frequent pattern accounts for less than 0.5% of all mapping patterns. A large number of patterns appear only once, which is reflected in the gradual ascension of the growth curve. This indicates that the language of the biomedical literature is very diverse, with a large number of different topics and writing styles.
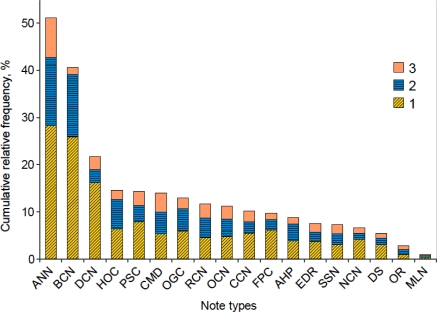
On the other hand, the growth curves of clinical notes rise sharply and flatten fairly fast. For example, the relative frequency of the most frequently occurring pattern in Ambulatory Nursing Notes accounts for 28% of all patterns of unambiguously mapped concepts; the top 3 patterns cover more than 50% of all instances of ST sequences within the analyzed corpus (Figure 2).
Figure 2.
Cumulative relative frequency of the top three patterns for each note type.
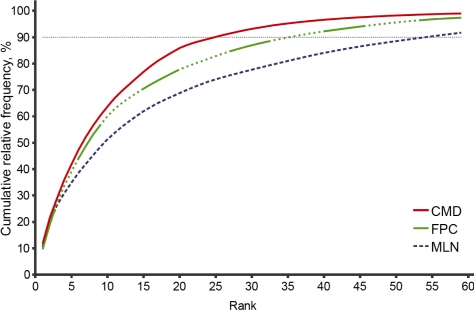
As Figure 3 demonstrates, clinical sublanguages use terms more narrowly than the language of MEDLINE abstracts. For example, far fewer semantic categories cover 90% of all mappings in clinical notes compared to MEDLINE abstracts.
Figure 3.
Semantic type growth curve: clinical notes versus MEDLINE abstracts.
Discussion
A sublanguage semantic schema (SSS) can be defined as a set of semantic types plus a set of rules that describe the co-occurrence of semantic types that account for a large proportion of all instances of terms appearing in the language of that domain. We have presented data showing that both the co-occurrence rule sets and the semantic type distributions vary widely across clinical note types, and vary from MEDLINE as well. These data support the proposition that clinical language is not homogenous and differs from biomedical language. Our findings suggest that clinical sublanguages can be modeled in a principled way. Once created, SSS can be used as a post-processing step to disambiguate terms that were mapped to multiple concepts that belong to different semantic types by selecting appropriate senses and discarding those meanings that are outside of the specific sublanguage grammar.
We have shown that instead of involving human experts to define the rules of semantic type relationships, these rules can be derived utilizing existing concept recognition tools, such as MetaMap, and existing semantic knowledge bases, such as UMLS. We are currently in the process of implementing and validating a system based on SSS approach.
Limitations
The current work makes several strong assumptions that will be tested in the future: that most unambiguous mappings produced by MetaMap are accurate; that inaccurate mappings produced by MetaMap result in a uniform distribution of semantic types that would not lead to strong co-occurrence patterns; and that the semantic type distributions in the notes that failed to process during MetaMap parsing follow the same distributions as in the notes that were processed successfully.
We also note that MetaMap failed to process a significant number of clinical texts. MetaMap assumes the well-composed sentence structure typical of MEDLINE abstracts. Clinical notes often lack that structure. In a related project we are investigating ways to pre-process clinical narratives such that concept extraction engines like MetaMap and MedLEE can process a wider variety of notes.
Conclusion
The purpose of the present research is to define an automatic method to acquire domain-specific rules that describe semantic sentence structure. Our results demonstrate that narratives created within different clinical domains exhibit the characteristics of sublanguages. A sublanguage semantic schema created using this approach potentially can be beneficial for word sense disambiguation. Implementation and validation of this approach is the next step of our ongoing research project.
Acknowledgments
This research has been supported by the NLM under grants T15LM007124 (fellowship), 5R21LM009967-02, and 3R21LM009967-01S1(ARRA). An allocation of computer time from the Center for High Performance Computing at the University of Utah is gratefully acknowledged.
References
- 1.Harris ZS. A Theory of Language and Information: A Mathematical Approach. Clarendon Press; 1991. [Google Scholar]
- 2.Stetson PD, Johnson SB, Scotch M, Hripcsak G. The sublanguage of cross-coverage. AMIA Annu Symp Proc. 2002:742–6. [PMC free article] [PubMed] [Google Scholar]
- 3.Rindflesch TC, Aronson AR. Ambiguity resolution while mapping free text to the UMLS Metathesaurus. Proceedings of the Symposium on Computer Applications in Medical Care. 1994:240–244. [PMC free article] [PubMed] [Google Scholar]
- 4.Friedman C. A broad-coverage natural language processing system. AMIA Annu Symp Proc. 2000:270–274. [PMC free article] [PubMed] [Google Scholar]
- 5.Leroy G, Rindflesch TC. Effects of information and machine learning algorithms on word sense disambiguation with small datasets. Int J Med Inform. 2005 Aug;74(7–8):573–585. doi: 10.1016/j.ijmedinf.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 6.Fan JW, Friedman C. Word sense disambiguation via semantic type classification. AMIA Annu Symp Proc. 2008:177–181. [PMC free article] [PubMed] [Google Scholar]
- 7.McCarthy D. Word Sense Disambiguation: An Overview. Language and Linguistics Compass. 2009;3(2):537–558. [Google Scholar]
- 8.Navigli R. Word sense disambiguation: A survey. ACM Comput Surv. 2009;41(2):1–69. [Google Scholar]
- 9.Pratt AW, Pacak MG. Proceedings of the 1969 conference on Computational linguistics. Morristown, NJ, USA: 1969. Automated processing of medical English; pp. 1–23. [Google Scholar]
- 10.Sager N, Lyman MS, Nhan NT, Tick LJ. Medical language processing: applications to patient data representation and automatic encoding. Methods Inf Med. 1995 Mar;34(1–2):140–146. [PubMed] [Google Scholar]
- 11.Aronson AR, Lang FM. The Evolution of MetaMap, a Concept Search Program for Biomedical Text. 2009.
- 12.MetaMap. 2009. http://metamap.nlm.nih.gov/
- 13.Aronson AR. Metamap: Mapping text to the UMLS Metathesaurus. Bethesda, MD: NLM, NIH, DHHS; 2006. [Google Scholar]