Abstract
Domain specific biomedical lexicons are extensively used by researchers for natural language processing tasks. Currently these lexicons are created manually by expert curators and there is a pressing need for automated methods to compile such lexicons. The Lexicon Builder Web service addresses this need and reduces the investment of time and effort involved in lexicon maintenance. The service has three components: Inclusion – selects one or several ontologies (or its branches) and includes preferred names and synonym terms; Exclusion – filters terms based on the term’s Medline frequency, syntactic type, UMLS semantic type and match with stopwords; Output – aggregates information, handles compression and output formats. Evaluation demonstrates that the service has high accuracy and runtime performance. It is currently being evaluated for several use cases to establish its utility in biomedical information processing tasks. The Lexicon Builder promotes collaboration, sharing and standardization of lexicons amongst researchers by automating the creation, maintainence and cross referencing of custom lexicons.
Introduction and background
The analysis of the enormous amount of publicly available biomedical data requires the use of biomedical ontologies to structure and annotate datasets with controlled terms in order to facilitate search, retrieval and data integration. Biomedical researchers routinely use ontologies and terminologies to annotate their data for better data integration and translational discoveries1. With the large number and variety (of formats and locations) of biomedical ontologies, the task of choosing the right ontology for an annotation task or for designing a curation tool is a challenge.
The National Center for Biomedical Ontology (NCBO) builds tools and services to assist the biomedical community in using ontologies to annotate and analyze biomedical data2, or to recommend an appropriate ontology for annotation3.
Increasingly, natural language processing (NLP) tools are used in annotation of biomedical data as well as in curation pipelines4. Even if the ontology to use in an NLP tool is identified and the tool can have programmatic access to a large number of biomedical ontologies in the NCBO BioPortal5, a significant amount of pre-processing is required to effectively use existing ontologies in natural language processing pipelines.
A lexicon(also called a dictionary) is a core component of any natural language processing system. For example, the SPECIALIST lexicon6 is a large syntactic lexicon of biomedical and general English. The use of lexicons, derived from terminologies and ontologies, for text mining and information extraction tasks is not new in the biomedical community. For example, the BioLexicon has been used in three text mining tasks a) BLTagger which is a dictionary-based parts-of-speech (POS) tagger; b) Enju full parser enriched using the lexicon; c) Lexicon-based query processing for information retrieval7. Medication information was extracted from discharge summaries using parsing rules written as a set of regular expressions and a user-configurable drug lexicon8. The authors acknowledge the necessity of careful lexicon selection for the extraction of drug information and to make the lexicon a configurable component in their system. The MedLEE lexicon was used to mine a clinical data warehouse for disease-finding associations9. The authors also mention that the MedLEE lexicon does not cover a large number of medical terms and using a larger coverage lexicon would improve the discovered associations. The authors acknowledge that an important class of named entity recognition approaches is lexicon-based and in order to improve the F-measure (combination of Precision and Recall) scores high-quality lexicons are essential10.
Basic text-mining resources, such as domain-specific thesauri and lexicons, need to be developed and shared across research groups and curation tasks; in order to extend the depth as well as breadth of the information that is curated, searched, and mined11. Ontologies and terminologies together with lexicons are important for advanced text mining and both are needed in order to produce highly accurate results needed by biomedical experts and to obtain broad coverage of biomedical text.12 The authors acknowledge that named entity recognition (NER) tasks require extensive domain-specific lexicons, which do not readily exist13. The authors argue that custom, domain specific lexicons are important background knowledge in medical language-processing systems14.
The main motivation for developing the NCBO Lexicon Builder Web service is to allow users to create custom domain-specific lexicons for specific NLP, data mining and information extraction tasks. For example, using our service, a researcher can compile a lexicon for identifying malignant skin tumors spanning multiple public ontologies. Currently, the creation of custom lexicons with biomedical ontology concepts is not a prevalent practice in the biomedical community for several possible reasons:
Creation of custom lexicons requires a huge investment and the accuracy and coverage of resulting lexicons is often questionable;
The large number of biomedical ontologies available for creating lexicons coupled with the frequent changes and overlap in these ontologies significantly increases the complexity;
Integrating related concepts over multiple related ontologies without the knowledge of the structure of ontologies is difficult and error prone; and limits the coverage of the lexicon.
The Lexicon Builder Web service automates the task of creating custom lexicons across multiple biomedical ontologies. The service leverages the Medline analysis15 to produce lexicons with high accuracy and coverage.
Methods
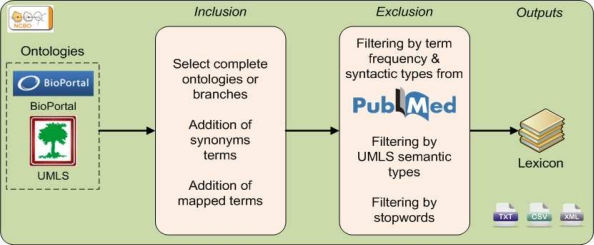
The workflow of the Lexicon Builder service is composed of two main steps (Figure 1) that define the inclusion criteria and exclusion criteria for ontology terms to pull into the lexicon. The output criteria allow the user to control the formatting of the output and ease the consumption of the service. First, the user’s input parameters are used to select the complete ontologies or ontology branches, followed by the addition of synonyms (if required) and related terms using the mappings among different ontologies present in BioPortal. The next step is the exclusion of term names that match certain stopwords, do not have the required syntactic types, exceed desired term frequency in Medline or do not have the required Unified Medical Language System (UMLS) semantic types. The final step is to filter information associated with the concepts that is not required in the output. Subsequently, the results are converted into a suitable format, compressed and returned to the user.
Figure 1.
Lexicon Builder service workflow.
The inclusion criteria in our system correspond to the following components.
An ontology component adds terms to the lexicon using one or more ontologies or a branch therein. For instance, if National Cancer Institute Thesaurus (NCI) Thesaurus and SNOMED-CT (Systematized Nomenclature of Medicine--Clinical Terms) are specified as ontologies, then the lexicon will include all terms within these two ontologies. The ontology component is parametrizable.
A concept hierarchy component adds terms to the lexicon using the parent-child hierarchy in the ontology. For instance, if the concept NCI/C0025202 (Melanoma in NCI Thesaurus) is specified as the parent term, the lexicon will include NCI/C0279693 (Intermediate Cell Type Uveal Melanoma in NCI Thesaurus) and NCI/C1334210 (Intermediate Cell Type Iris Melanoma in NCI Thesaurus) which are child nodes of Melanoma. The parent or ancestor concept for specifying the branch of the ontology is configurable.
A synonym-expansion component adds all the synonyms for a term in addition to the preferred name for the term. For example, if the term NCI/C0278884 (Recurrent Melanoma in NCI Thesaurus) is added to the lexicon, this component will add new terms (such as Recurrent Malignant Skin Melanoma, Recurrent Malignant Melanoma of Skin, Recurrent Cutaneous Melanoma) to the lexicon in addition to the preferred name (Recurrent Melanoma of the Skin). The use of synonyms is optional.
An ontology-mapping component adds new terms based on existing one-to-one mappings among different ontologies16. For example, if the term NCI/C0025202 (melanoma in NCI Thesaurus) is added to the lexicon in the first phase, this component can add 38865/DOID:1909 (Naevocarcinoma in Human Disease), defined as a synonym of melanoma in Human Disease, but not in NCI Thesaurus. The type of mapping to use is parametrizable. Note that constraints specified by the ontology component and concept hierarchy component do not apply to terms retrieved using the mapping component.
The exclusion criteria in our system correspond to the following components;
A medline-count component excludes terms based on the frequency of their occurrence in Medline abstracts. For example, if a Medline term frequency of 10000 is specified as the cutoff, then the concept NCI/C1883030 (Signet-Ring Melanoma in NCI Thesaurus) is included in the lexicon, whereas the concept NCI/C0012634 (Disease in NCI Thesaurus) is excluded from the lexicon since the term frequency for NCI/C0012634 is greater than 10000. The authors argue that the term frequency is an appropriate way to filter out common concepts, which are likely to be uninformative for NER or information retrieval15.
A syntactic-type component retains terms based on the predominant syntactic type of the term over all of Medline sentences. For instance, if the desired syntactic type is Noun Phrase then the concept NCI/C1709220 (Neurotropic Melanoma in NCI Thesaurus) is retained in the lexicon, whereas the concept NCI/C0855030 (Stage I Superficial Spreading Melanoma in NCI Thesaurus) is excluded since the predominant syntactic type of the terms for NCI/C0855030 is not of type Noun Phrase.
A semantic type component filters concepts based on existing UMLS semantic types. For example, in order to keep those child terms under the hierarchy rooted at NCI/C0481391 (behavior-related disorder in NCI Thesaurus) which are of semantic type Neoplastic Process, the user can specify NCI/C0481391 as the parent concept and the semantic type T191(Neoplastic Process). The semantic component will keep child terms such as NCI/C1332228 (Alcohol-Related Hepatocellular Carcinoma) which has semantic type Neoplastic Process and eliminate child terms such as NCI/C1335368 (Passive-Aggressive Behavior) which has the semantic type of Mental or Behavioral Dysfunction from the lexicon.
The stopwords component excludes terms if they are contained in the specified (user-defined or system default) stopwords. For example, if the concept NCI/C0025202 (melanoma in NCI Thesaurus) is specified as parent concept and ‘Stage I’ as the stopword, then the concept NCI/C0855030 (Stage I Superficial Spreading Melanoma in NCI Thesaurus) is excluded form the lexicon. The stopwords to use are parametrizable.
The output criteria in our system ease the integration of our output into user workflows.
An output fields component allows the user to specify the information (such as the term id, the term Uniform Resource Identifier (URI), the ids of the ontologies accessed) to include in the lexicon for every term. This is useful for maintainence of the lexicon over time. An output format component generates the lexicon in several formats. The currently supported formats include text, tab delimited and Extensible Markup Language(XML) formats in both compressed or uncompressed versions.
The Lexicon Builder Web service aims to address the issues (mentioned in the Introduction section)8,9,10 and enables users to follow the recommendation on sharing lexicons11. The Web service can produce and maintain lexicons with minimal amount of time and effort, eases integration into other workflows. Creating a new lexicon requires a few iterations where in each iteration the user specifies the parameters, retrieves the lexicon, analyzes the output for adequacy and subsequently changes the parameters in order to get the desired lexicon.
Results
Prototype implementation
We implemented the service using, all the (English) ontologies in UMLS and a subset of the NCBO BioPortal ontologies. These ontologies offer a dictionary of 4,222,921 concepts and 7,943,757 terms. Concepts are identified by either the UMLS Concept Unique Identifier (CUI) for identifying ULMS concepts/senses or NCBO Uniform Resource Identifier (URI) for identifying BioPortal concepts. By the time the LexiconBuilder prototype was implemented, the UMLS ontologies were not imported in BioPortal. In future versions, only BioPortal URIs will be used. In the second step of the workflow, our system currently uses the UMLS metathesaurus CUI-based and user-defined mappings in BioPortal to expand the lexicon produced by the first step of the workflow. The Lexicon Builder is publicly available(http://labs.bioontology.org/LexiconBuilder) and is deployed only as a RESTful (REpresentational State Transfer) Web service.
Evaluation
The ideal way to evaluate our results would be to compare both coverage and accuracy with manually created lexicons for the same domain. However, such an evaluation is very difficult in practice, since manually creating a large number of lexicons for evaluation requires time and effort, expert curators and/or end users who have considerable domain knowledge and are willing to evaluate the system.
We evaluated our system for technical accuracy by manually cross-checking terms in the generated lexicons with ontology terms present in BioPortal. We conducted the evaluation by creating lexicons from ontologies of differing sizes and formats. The ontologies and their corresponding formats, the parent concept in the ontology hierarchy whose descendents we verified, and the number of unique concepts from the hierarchy that were pulled into the lexicon are given below (Table 1). In each case our service pulled out all the desired concepts in the appropriate sub-branches without any unwanted concepts.
Table 1.
Verification statistics on BioPortal ontologies
| Ontology | Format | Parent Concept | Unique Concepts |
|---|---|---|---|
| Cell Type | OBO | Fungal cell | 17 |
| SNOMED-CT | RRF | Subclass Eucestoda (organism) | 229 |
| NCI Thesaurus | OWL | Bacterial Infection | 86 |
We also evaluated the performance of our system on a set of five ontologies of widely different sizes. The time taken to generate the lexicons along with the lexicon size (uncompressed) and number of unique concepts present in the lexicon are given below (Table 2).
Table 2.
Performance statistics on BioPortal ontologies
| Ontology | Time-minutes | Uncompressed Size(MB) | Unique Concepts |
|---|---|---|---|
| Cell Type | 1 | 4 | 609 |
| SNOMED-CT | 15 | 4198 | 262372 |
| Human Disease | 1 | 67 | 10193 |
| NCI Thesaurus | 2 | 338 | 32120 |
| Drosophila Gross Anatomy | 1 | 4 | 917 |
Novel features of our Web service
With the large number of ontologies present in BioPortal, mappings between ontology terms can identify and retrieve relevant concepts for other ontologies that are unfamiliar to the user. Lexicons generated using the inter-ontology mappings facilitate the integration of datasets that are annotated with differing ontologies.
Users can use the BioPortal visualization service in conjunction with the Lexicon Builder service to visualize and understand the ontology structure (hierarchies), subsequently change the configurable components of the Lexicon Builder service to refine the retrieved lexicon. Visualization can be especially helpful for generating large lexicons from multiple ontology branches.
Our service also addresses the issue of version management and cross-referencing of lexicons. When changes occur to the underlying ontologies, new terms are added to the ontologies or existing terms are changed or deleted, the users can simply retrieve a newer version of their lexicon using the parameters used before. Ontologies in BioPortal have URIs(uniform resource identifiers) for all ontology terms. Researchers can cross-reference lexicons generated using our service in a straightforward manner since the terms will have the same unique identifiers across all custom lexicons. Moreover, custom-built terminologies are mapped to other terminologies when uploaded into BioPortal. Hence, they can be expanded using the mappings.
Use cases
The lexicons generated using our Web service can be utilized in several applications in the biomedical community, many of which provided the driving usecases for our work. The created lexicons can be used for domain or requirement specific text annotation tasks. e.g. the annotation of protein mutations with disease terms17 They can be instrumental for ontology related tasks such as ontology learning, ontology enrichment and information extraction. Another application where these lexicons can be used is for the purpose of tagging entities in web pages for enhancing the user browsing experience18 in the Reflect project. Just as it is possible to “reflect” a protein name, it is possible tag disease entities to add context specific information using a cancer lexicon generated using our system.
The service is currently being evaluated for use in external workflows. (1) Researchers at the University of Pittsburgh School of Medicine developing the Ontology Development and Information Extraction (ODIE) toolkit (http://www.bioontology.org/ODIE-project)are evaluating the service for creating workflow specific lexicons such as a ‘neoplastic skin disorders’ lexicon to use in ODIE workflows for concept recognition tasks. (2) The BLULab (http://www.dbmi.pitt.edu/blulab/) research group at the University of Pittsburgh School of Medicine is researching semi-automated, data-driven methods for extending Topaz’s lexicon in the syndromic surveillance ontology and is evaluating our service as one of the methods to be used with their system. (3) Researchers at the European Molecular Biology Laboratory working on the Reflect project (http://reflect.ws/) are evaluating the Lexicon Builder for compiling specific lexicons, such as for human diseases, for use in their work.
Discussion and Related work
The SPECIALIST lexicon6 provides the lexical information needed for the SPECIALIST Natural Language Processing System. It is intended to be a general English lexicon that includes many biomedical terms. Coverage includes both commonly occurring English words and biomedical vocabulary discovered in the National Library of Medicine (NLM) Test Collection and the UMLS Metathesaurus. The BioLexicon19, aggregates terms from project partners. Term entries in the BioLexicon are linked to an ontology and function as the terminological resource for extracting information from documents. The lexicon used in their work is compiled from online dictionaries and suggestions from professional bio-medical researchers20. The domain-specific lexicon can be edited to meet the user’s needs, and hence, it can be expanded, when required.
We argue that our service provides functionality not provided by the systems discussed above.
Our system produces lexicons using all the (English) UMLS terminologies as well as the Open Biological and Biomedical Ontologies(OBO) and other formats in BioPortal. The SPECIALIST lexicon is an English lexicon with also many biomedical terms. As a result, it lacks the breadth of coverage of biomedical concepts.
The Lexicon Builder uses BioPortal which has the largest set of publicly available ontologies as compared to any other existing repository till date. In contrast, BioLexicon relies on partner data to create the lexicons. In our case, the number of ontologies that can be used to generate the lexicons, in conjunction with the ability to create and maintain custom lexicons when the underlying ontologies change, is a significant advance over BioLexicon.
The system proposed20 is very similar to the BioLexicon system with respect to the data sources used to create the lexicons. Thus, their system is susceptible to the same limitations as BioLexicon.
The UMLS MetamorphoSys tool allows users to create customized UMLS Metathesaurus subsets. However, it doesn’t work with OBO ontologies. The Lexicon Builder gives more fine grained control, by allowing to choose ontology branches, concept hierarchy, filter by stopwords, syntactic types and Medline counts in addition to filters common to both tools such as semantic types. It supports the XML format and can be easily consumed as a Web service.
The Lexicon Builder system distinguishes itself from previous efforts for several reasons:
Available as a Web service that can be integrated in existing workflows, enabling easy sharing, adoption and cross-referencing of lexicons.
Uses publicly available ontologies instead of third-party or partner data. This ensures better coverage and requires less investment of time and effort to maintain lexicons.
Has access to the largest available set of biomedical ontologies from the UMLS Metathesaurus and NCBO BioPortal as well as access to ontology visualization tools that simplify the task of lexicon parameter selection.
Conclusion
Building lexicons from biomedical ontologies is crucial for text-mining and natural language processing tasks. We have presented a web service for ontology-based generation of lexicons from BioPortal.
Our Lexicon Builder service has access to ontologies spanning both UMLS and Open Biomedical ontologies. The service leverages the structural information present in ontologies as well as the syntactic type information on individual terms mined from Medline.
The service can be customized to a user needs (in terms of parameters and biomedical ontologies used) and is being evaluated by the biomedical community for its utility in creating custom lexicons.
Acknowledgments
This work is supported by NIH grant U54 HG004028 in support of the National Center for Biomedical Ontology, one of the National Centers for Biomedical Computing.
References
- 1.Shah NH, Jonquet C, Chiang AP, Butte AJ, Chen R, Musen MA. Ontology-driven indexing of public datasets for translational bioinformatics. BMC Bioinformatics. 2009;10(Suppl 2):S1. doi: 10.1186/1471-2105-10-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jonquet C, Shah NH, Musen MA. The Open Biomedical Annotator, AMIA Summit on Translational Bioinformatics. 2009:56–60. [PMC free article] [PubMed] [Google Scholar]
- 3.Jonquet C, Musen MA, Shah NH. Building a Biomedical Ontology Recommender Web Service. Biomedical Semantics. 2010;1(S1) doi: 10.1186/2041-1480-1-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dowell KG, McAndrews-Hill MS, Hill DP, Drabkin HJ, Blake JA. Database (Oxford) 2009. Integrating text mining into the MGI biocuration workflow. 2009:bap019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noy NF, Shah NH, Whetzel PL, et al. BioPortal: ontologies and integrated data resources at the click of a mouse. Nucleic Acids Research. 2009;37:170–173. doi: 10.1093/nar/gkp440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Browne AC, McCray AT, Srinivasan S. The Specialist Lexicon. Technical Report. 2000 [Google Scholar]
- 7.Sasaki Y, Thompson P, McNaught J, Ananiadou S. Three BioNLP Tools Powered by a Biological Lexicon. In Proc of the Demonstrations Session EACL. 2009:61–64. [Google Scholar]
- 8.Gold S, Elhadad N, Xinxin Z, Cimino JJ, Hripcsak G. Extracting Structured Medication Event Information from Discharge Summaries. In Proc of AMIA Annual Symp 2008. 2008:237–41. [PMC free article] [PubMed] [Google Scholar]
- 9.Cao H, Markatou M, Melton GB, Chiang MF, Hripcsak G. Language Processing and its future in medicine: Mining a clinical data warehouse to discover disease-finding associations using co-occurrence statistics. Proc of AMIA Annual Symp 2005. 2005:106–110. [PMC free article] [PubMed] [Google Scholar]
- 10.Cohen AM, Hersh WR. A survey of current work in biomedical text mining. Briefings in Bioinformatics. 2005;6(1):57–7. doi: 10.1093/bib/6.1.57. [DOI] [PubMed] [Google Scholar]
- 11.Altman RB, Bergman CM, Blake J, et al. Text mining for biology - the way forward: opinions from leading scientists. Genome Biology. 2008;9(Suppl 2):S7. doi: 10.1186/gb-2008-9-s2-s7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Spasic I, Ananiadou S, McNaught J, Kumar A. Text mining and ontologies in biomedicine: Making sense of raw text. Briefings in Bioinformatics. 2005;6(3):239–251. doi: 10.1093/bib/6.3.239. [DOI] [PubMed] [Google Scholar]
- 13.Yang J, McDonald RT, Lerman K, et al. Automated recognition of malignancy mentions in biomedical literature. BMC Bioinformatics. 2006;7:492. doi: 10.1186/1471-2105-7-492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Xu R, Supekar K, Morgan A, Das A, Garber A. Unsupervised Method for Automatic Construction of a Disease Dictionary from a Large Free Text Collection. In Proc of AMIA Annual Symp. 20082008:820–824. [PMC free article] [PubMed] [Google Scholar]
- 15.Xu R, Musen MA, Shah NH. A Comprehensive Analysis of UMLS Metathesaurus Terms Using Eighteen Million MEDLINE Abstracts. Proc of AMIA Annual Symp 2010. 2010 [PMC free article] [PubMed] [Google Scholar]
- 16.Ghazvinian A, Noy NF, Musen MA. Creating Mappings For Ontologies in Biomedicine: Simple Methods Work, In Proc of AMIA Annual Symp 2009. 2009 [PMC free article] [PubMed] [Google Scholar]
- 17.Hancock D, Morrison N, Velarde G, Field D. An interactive web-based tool for the automated detection of ontological terms in unstructured, free-text annotation. Nature Precedings. 2009 [Google Scholar]
- 18.Pafilis E, O’Donoghue SI, Jensen LJ, et al. Reflect: Augmented Browsing for the Life Scientist. Nature Biotechnology. 2009;27:508–510. doi: 10.1038/nbt0609-508. [DOI] [PubMed] [Google Scholar]
- 19.Quochi V, Monachini M, Gratta RD, Calzolari N. A Lexicon for Biology and Bioinformatics: TheBootStrep Experience. In Proc of the Sixth International Language Resources and Evaluation 2008 [Google Scholar]
- 20.Chiang JH, Yu HC, Hsu HJ. GIS: a biomedical text-mining system for gene information discovery. Bioinformatics. 2004;20(1):120–1. doi: 10.1093/bioinformatics/btg369. [DOI] [PubMed] [Google Scholar]