Abstract
Objectives:
To assess whether 1) the necessary drug classes and 2) the necessary drug-class membership relations are represented in biomedical terminologies in order to support clinical decision regarding drug-drug interactions.
Methods:
In order to investigate drug classes and drug-class membership in clinical terminologies, we start by establishing a reference list of these entities. Then, we map drugs and classes to the UMLS, where we investigate their relations.
Results:
186 (83%) of the 223 names for drug classes mapped to the UMLS. The single best source is SNOMED CT with 75%. 140 (89%) of the 157 drug-membership relations were found in the UMLS.
Conclusions:
One important category of drug classes missing from all clinical terminologies is related to drug metabolism by the Cytochrome P450 enzyme family.
Introduction
The average number of prescription drugs purchased per capita in the United States exceeds 12 [1]. The mortality risks for even individually prescribed drugs are roughly equal to or exceed the corresponding risk for passenger vehicle travel (11 per 100,000 person-years) [2]. Many drugs are prescribed concomitantly, which increases the risk of adverse drug events [3]. Most existing computerized provider order entry (CPOE) systems present alerts for known drug-drug interactions which, if selected carefully for severe interactions, lead to changes in the patient’s medication orders [4–5]. Although the influence of clinical decision support (CDS) provided by CPOE systems on reduction of prescription errors has been studied for over ten years, a recent review concluded that “the role of decision support in minimizing severe prescribing error rates requires investigation” [6]. Prescription errors reported in the 2006 IOM report on medication errors ranged from 12.3 to 1,400 errors per 1,000 hospital admissions [7]. The vision for optimal, CDS-enabled medication management outlined by the Agency for Healthcare Research and Quality (AHRQ) accentuates the importance of standarized and readily available representation of drug information (addressing dosing, side effects, costs, interactions, etc) during prescriber ordering and verification of the order by pharmacy staff, listing double-checking for interactions as the first item in the drug verification and dispensing process [8].
Authoritative information about drug interactions is summarized in the drug package inserts maintained by the Food and Drug Administration (FDA) and available at the National Library of Medicine (NLM) DailyMed website [9]. This information however only partially addresses the AHRQ request for availability of widely used, standardized, and practical formats for expressing medication-specific information in both human and machine-readable form. Namely, this information is only human readable and not standardized.
Standardized, actionable knowledge about drug-drug interactions is available from proprietary sources (e.g., First DataBank), as well as from a limited number of publicly-available sources (e.g., NDF-RT). However, the generic rules found in these systems may need to be adapted to the specificity of a local practice or hospital. Clinical decision support (CDS) rules related to drug-drug interactions are generally expressed in terms of relations between an individual drug and a therapeutic or pharmacologic class (e.g., between itraconazole and H2-receptor antagonists) or between classes of drugs (e.g., between ACE Inhibitors and Potassium supplements). While drug-drug interactions themselves are not expected to be represented in clinical terminologies, the vocabulary for expressing such interactions (i.e., drug and class names, and drug-class membership relations) should be present in these terminologies. Drugs provide entry points into the electronic health records (EHR) system, drugs and classes are referenced in the CDS, and drug-class membership relations enable the link between EHR and CDS.
The objective of this study is to investigate the degree to which biomedical terminologies provide adequate support for clinical decision related to drug-drug interactions. More specifically, we assess whether 1) the necessary drug classes and 2) the necessary drug-class membership relations are represented in biomedical terminologies. In this preliminary investigation, we focus on a limited set of drugs frequently involved in severe interactions with other drugs.
Background
The relation between individual drugs and drug classes is generally represented through a taxonomic relation (isa) in biomedical terminologies. For example, the relation between Omeprazole and Proton pump inhibitors is represented as an isa relation in SNOMED CT, MEDCIN and the NCI Thesaurus. In some cases, an associative relationship (specified or not) is used between the drug and the class. This is the case, for example, between candesartan and Angiotensin II receptor antagonist in USPMG.
In other terminologies, the relation is more complex. In NDF-RT, for example, there are no explicit relations between ingredients from the drug hierarchy and classes from the External Pharmacologic Classes hierarchy. However, NDF-RT uses description logics (DL) for its representation and, using a DL reasoner, it is possible to make inferences. In particular, it is possible to compute relations between drugs and classes based on the properties asserted for these concepts. Such inferences assume that the drug classes are defined classes, which is currently not the case. However, after modifying the definitions slightly, it is possible to reclassify the knowledge base and to obtain drug-class relations [10].
Materials
UMLS
The Unified Medical Language System® (UMLS®) is a terminology integration system developed at the National Library of Medicine. The UMLS Metathesaurus® integrates almost 150 biomedical vocabularies. Synonymous terms from the various source vocabularies are grouped into one concept. Additionally, the Metathesaurus records the relations asserted among terms in the source vocabularies, including hierarchical, associative and mapping relations. Version 2009AB of the UMLS is used in this study. This version contains approximately 2.1M concepts and 40M relations.
Methods
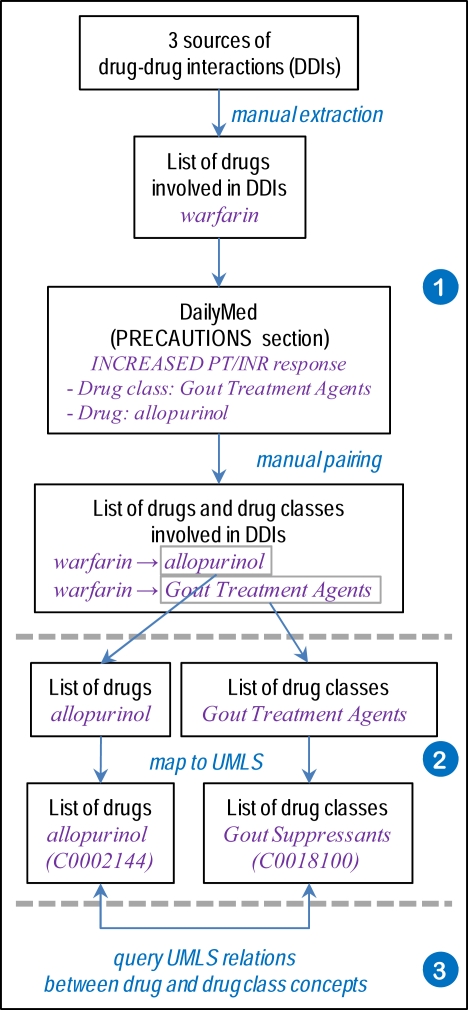
In order to investigate drug classes and drug-class membership, as shown in Figure 1, we start by establishing a reference list of these entities (1). Then, we map drugs and classes to the UMLS (2), where we investigate their relations (3).
Figure 1.
Overview of the methods.
Establishing a reference list of drug classes and drug membership for these classes
We started by establishing a list of drugs of interest, in order to seed a list of drug classes known to interact with these drugs. Finally, we collected drug membership information for these drug classes from an authoritative source.
Establishing a list of drugs of interest
We compiled a list of drugs frequently causing drug-drug interactions from the following three sources.
Beers’ list of medications inappropriate for use in elderly patients [11]. This list contains 66 drugs known to have severe adverse outcomes.
Drugs listed in the “Top Ten Dangerous Drug Interactions in Long-Term Care” maintained by the Medication Management Project [12].
The Cytochrome P450 Drug Interaction table, a reference list of drugs whose metabolism is mediated by various members of the CYP450 enzyme family [13].
From these lists, we selected only individual drugs (not drug classes). Examples of such drugs include alprazolam (from Beer’s and the CYP450 lists), warfarin (from the Top Ten and CYP450 lists) and amiodarone (common to the three lists).
Establishing a list of drug classes involved in drug-drug interactions
Starting from the individual drugs in the list we just established, we used FDA’s Structured Product Labels, an authoritative source of drug information, for identifying classes of drugs known to interact with the drugs of interest. More specifically, we used DailyMed [9] to acquire the labels as XML documents, from which we parsed the “Precautions” section semi-automatically in order to extract all drug classes mentioned. Examples of drug classes associated with warfarin include Antiandrogen, Gout Treatment Agents and Steroids, Adrenocortical.
Establishing reference drug-class membership information
In addition to the drug classes listed in the Structured Product Labels (SPLs), we also collected the names of individual drugs. We paired drugs and classes manually, when the association was not made explicit through the discourse structure of the label (e.g., when an individual drug is cited as a representative example of a drug class). For example, from the SPL information for warfarin, the individual drug allopurinol was associated with the class Gout Treatment Agents.
Mapping drug classes to clinical terminologies
The list of drug classes was mapped to the UMLS through exact match or after normalization. Multiple mappings were disambiguated using the semantic types. We attempted to map the drug classes manually, when no automated match was found. All mappings, automated and manual, were reviewed by the authors. Based on the mapping to UMLS, we traced each drug class back to specific source vocabularies in the UMLS in order to assess the coverage provided by each source. Analogously, we mapped each individual drug identified as the representative of a class to UMLS concepts automatically.
Identifying drug-class membership
With individual drugs and classes both mapped to UMLS concepts, we explored the following UMLS relations in order to find a link between the drug and its class. We first looked for the class concept in the direct ancestors (parent of broader concepts) of the drug concept. We then used the transitive closure of such hierarchical relations, allowing the class to be in indirect hierarchical relation to the drug. Finally, we also explored the (direct) associative (i.e., non-hierarchical) relations of the drug and searched for the class among the related concepts.
Results
Reference list of drug classes and drug membership for these classes
List of drug classes involved in drug-drug interactions
From the 56 names of individual drugs extracted from the three sources, 40 were selected because they had an entry in DailyMed and were mentioned as being involved in drug interaction in the text of the label. From these 40 individual drugs, we harvested 223 names for drug classes, including minor variants (e.g., ACE Inhibitors vs. ACE-inhibitors), but excluding three misspelled drug class names.
Reference drug-class membership information
A total of 161 reference drug-class pairs were extracted largely from the tables of individual drugs and classes listed in SPL information for warfarin. All but four drug names were mapped to the UMLS, resulting into 157 unique concepts, each of which was associated with one class.
Drug classes in the UMLS
Of the 233 names for drug classes, 134 (60%) mapped automatically to UMLS concepts, including Cardiac glycosides (C0007158) and Anti-retroviral Agents (C0599685). In 7 cases, the mapping was ambiguous (to two concepts), but both concepts were deemed valid mappings in the context (e.g., the mapping of Macrolides to both Macrolides (C0282563) and Macrolide Antibiotics (C0003240)).
Of the 89 classes without automatic mapping, 52 could be mapped manually to a UMLS concept. Causes for mapping failure included missing synonyms (22 cases, e.g., Tuberculosis Agents for Anti-tubercular Agents (C0003448)); overly specified classes (e.g., 16 cases, e.g., Oral Contraceptives, Estrogen Containing mapped to Contraceptives, Oral (C0009905)); close, but slightly different concepts (14 cases, e.g., coumarin-type anticoagulants mapped to Anticoagulants, Oral (C0354604)).
A total of 37 names (17%) remained unmapped, because the corresponding concept is missing from the UMLS. A vast majority (31) of these classes refer to the metabolism of the drugs, more specifically to a particular enzyme of the CYP450 family, e.g., Cytochrome P450 Inducers, CYP2C8 inhibitors, and CYP3A4 substrates. The remaining unmapped classes refer to side effects (e.g., drugs known to prolong the QTc interval, Hepatotoxic Drugs) or to a mix of chemical and physiological properties (e.g., indirectacting amines).
Overall, 186 (83%) of the 223 names for drug classes mapped to the UMLS. A total of 134 distinct UMLS concepts were mapped to, including the multiple mappings for 7 drug names mentioned earlier.
Drug classes in specific clinical terminologies
By design, each of our 134 drug classes we selected is present in at least one source vocabulary from the UMLS. SNOMED CT covers 101 (75%) of the drug classes, followed by MeSH (72%). SNOMED International, CRISP, the Read Codes, the Alcohol and Other Drugs Thesaurus, MEDCIN and NDF-RT all cover at least 50% of these drug classes.
From the perspective of drug classes, 17% of the classes are represented in 15 or more sources (excluding translations), 41% in 10–14 sources, 25% in 5–9 sources, and 25% in 1–4 sources.
Drug-class membership in clinical terminologies
Overall, 140 (89%) of the 157 drug-membership relations were found in the UMLS. The following types of relations were found between the drug and its class. In 139 cases (89%), the class is an ancestor of the drug (direct ancestor in 124 cases). Examples of direct relations include Cefotetan--Cephalosporins and Methimazole--Antithyroid Drugs, while ezetimibe--Hypolipidemics is found only indirectly through the concept Antilipemic agents. In 17 cases, the drug and the class are in associative relation, but in all but one case (Doxazosin--Central Alpha1-Blockers), the associative relation coexists with a hierarchical relation. Finally, no relation is found between the drug and the class in 17 cases (11%), including Cholestyramine Resin--Bile Acid-Binding Resins, olsalazine--Ulcerative Colitis Agents, and Ranitidine--Gastric Acidity and Peptic Ulcer Agents.
Extended example
Consider a situation when a patient taking amiodarone needs a broad-spectrum antibiotic. The patient’s physician intends to prescribe ciprofloxacin using the CPOE system. The clinical decision support system connected to CPOE might store (in structured, actionable form) the official recommendations for amiodarone: “There have been reports of QTc prolongation, with or without TdP, in patients taking amiodarone when fluoroquinolones, macrolide antibiotics, or azoles were administered concomitantly.” Using the NDF-RT, the CDS system will determine that Ciprofloxacin (C0008809) is a Fluoroquinolones (C291546) and issue an alert. Ideally the system should also recognize that the other class to which ciprofloxacin belongs is a broad-spectrum antibiotic and suggest a representative that does not belong to antibiotic subclasses that cause prolonged QT. However, the class broad-spectrum antibiotics is not represented as class in the UMLS.
Discussion
Missing classes
Overall, the coverage of drug classes in the UMLS is relatively good. Because the UMLS draws terms from a wide variety of clinical and other vocabularies, we had expected the coverage to be greater. In fact, missing from the UMLS, i.e., not represented in any of its source vocabularies, is one particular type of drug class: classes defined in reference to drug metabolism. As we have gained knowledge about the functions of the subfractions of the Cytochrome P450 enzyme family over the past decade, many drugs have been classified as inducers, inhibitors and substrates of this enzymatic system. On the bedside, this knowledge translates into recommendations about the use of combinations of drugs affecting one particular enzyme. Interestingly, the enzymes themselves are represented in several clinical terminologies (e.g., CYP2C8 in SNOMED CT and NDF-RT). Moreover, NDF-RT defines a relation (metabolizes) between some drugs (e.g., omeprazole) and CYP2C8. However, this knowledge would need to be further processed in order to infer a class of drugs metabolized by CYP2C8.
At the level of individual vocabularies, the best coverage of drug classes is 75% (for SNOMED CT). It is difficult to say, however, whether SNOMED CT should increase its coverage of drug classes or if the editorial guidelines for the Structured Product Labels should prescribe greater standardization and mandate the use of a reference terminology in order to reduce the variability of names for drug classes. The coverage provided by NDF-RT was disappointing to us, since this vocabulary provides a large hierarchy of so-called External Pharmacologic Classes. This finding requires further investigation.
Drug-class membership relations
Overall, the coverage of the drug-class membership relations was good (89%). However, by design, the coverage was inspected only for those classes found in the UMLS. In practice, based on this sample, the classes found in the UMLS are generally appropriately linked to their drug representatives in clinical terminologies. This coverage also represents a “best case scenario”, as relations from any terminology in the UMLS were allowed.
The fact that the most common type of relation between a drug and a class is isa, reflects that individual drugs and classes are generally part of the same hierarchies. On the one hand, this feature makes it easy for users to navigate between drugs and classes. On the other, it probably means there are few distinctive elements that would allow an agent (a computer) to distinguish between them automatically. One exception is NDF-RT in which additional properties clearly identify ingredients, clinical drugs and classes.
Lack of standardization in the names of the drug classes may lead to what appears as false negatives. For example, from the DailyMed tables, we associated Spironolactone with the class Adrenal Cortical Steroid Inhibitors. In the UMLS, the class for Spironolactone is Aldosterone Antagonists. While releated in meaning, the two classes do not share any hierarchical relations and the class found in DailyMed is not corroborated by UMLS relations. Other examples include Bile Acid-Binding Resins vs. Bile acid sequestrant antilipemic agent.
Limitations and future work
This study did not intend to be exhaustive, or even representative of any dataset. Therefore, any findings must be generalized with great caution. In future work, we are planning a comprehensive study of the representation of drug classes and drug-class membership in biomedical terminologies.
Using DailyMed as our reference provided us with an authoritative source, but imposed us to parse it mostly manually in order to establish our reference. Document formatting templates are no substitute for structured, standardized and coded information. We are also working on the annotation of the Structured Product Labels with reference terminologies.
Finally, this study probably does not fully do justice to the wealth of information present in NDF-RT. However, as mentioned earlier, we found this information difficult to extract (e.g., when the ontology needs to be reclassified after modifying some definitions in order to compute the necessary inferences).
Acknowledgments
This research was supported in part by the Intramural Research Program of the National Institutes of Health (NIH), National Library of Medicine (NLM).
References
- 1.Data source: Retail prescription drugs filled at pharmacies (Annual per capita): http://statehealthfacts.org
- 2.Cohen JT, Neumann PJ. What’s more dangerous, your aspirin or your car? Thinking rationally about drug risks (and benefits) Health Aff (Millwood) 2007;26(3):636–46. doi: 10.1377/hlthaff.26.3.636. [DOI] [PubMed] [Google Scholar]
- 3.Halkin H, Katzir I, Kurman I, Jan J, Malkin BB. Preventing drug interactions by online prescription screening in community pharmacies and medical practices. Clin Pharmacol Ther. 2001;69(4):260–5. doi: 10.1067/mcp.2001.114228. [DOI] [PubMed] [Google Scholar]
- 4.Kuperman GJ, Bobb A, Payne TH, Avery AJ, Gandhi TK, Burns G, et al. Medication-related clinical decision support in computerized provider order entry systems: a review. J Am Med Inform Assoc. 2007;14(1):29–40. doi: 10.1197/jamia.M2170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paterno MD, Maviglia SM, Gorman PN, Seger DL, Yoshida E, Seger AC, et al. Tiering drug-drug interaction alerts by severity increases compliance rates. J Am Med Inform Assoc. 2009;16(1):40–6. doi: 10.1197/jamia.M2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reckmann MH, Westbrook JI, Koh Y, Lo C, Day RO. Does computerized provider order entry reduce prescribing errors for hospital inpatients? A systematic review. J Am Med Inform Assoc. 2009;16(5):613–23. doi: 10.1197/jamia.M3050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Institute of Medicine; 2006. Preventing medication errors: Quality chasm series. http://www.iom.edu/Reports/2006/Preventing-Medication-Errors-Quality-Chasm-Series.aspx. [PubMed] [Google Scholar]
- 8.Agency for Healthcare Research and Quality A vision for optimal, CDS-enabled medication management. 2009. http://healthit.ahrq.gov/images/mar09_cds_book_chapter/CDS_MedMgmnt_ch_1_sec_7_vision_med_mgmnt.htm.
- 9.DailyMed: http://dailymed.nlm.nih.gov/
- 10.Bodenreider O, Mougin F, Burgun A. Automatic determination of anticoagulation status with NDF-RT. Proceedings of the 13th ISMB’2010 SIG meeting “Bio-ontologies”; 2010; pp. 140–143. [Google Scholar]
- 11.Fick DM, Cooper JW, Wade WE, Waller JL, Maclean JR, Beers MH. Updating the Beers criteria for potentially inappropriate medication use in older adults: results of a US consensus panel of experts. Arch Intern Med. 2003;163(22):2716–24. doi: 10.1001/archinte.163.22.2716. [DOI] [PubMed] [Google Scholar]
- 12.Top ten dangerous drug interactions in long-term care: http://www.scoup.net/m3project/topten/
- 13.Drug Interactions: Cytochrome P450 drug interaction table. Indiana University School of Medicine: http://medicine.iupui.edu/clinpharm/ddis/table.asp.