Abstract
Cause of death data is an invaluable resource for shaping our understanding of population health. Mortality statistics is one of the principal sources of health information and in many countries the most reliable source of health data. 1 A quick classification process for this data can significantly improve public health efforts. Currently, cause of death data is captured in unstructured form requiring months to process. We think this process can be automated, at least partially, using simple statistical Natural Language Processing, NLP, techniques and the Unified Medical Language System, UMLS, as a vocabulary resource. A system, Medical Match Master, MMM, was built to exercise this theory. We evaluate this simple NLP approach in the classification of causes of death. This technique performed well if we engaged the use of a large biomedical vocabulary and applied certain syntactic maneuvers made possible by textual relationships within the vocabulary.
INTRODUCTION
Death certificates have been an important source of information on population health since the early 12th century. As early as 1385, cities in Northern Italy, weary of plague and its attendant financial consequences, began registering deaths and recording causes of death to facilitate surveillance. The Bills of Mortality of Florence and Mantua’s “Books of the Deceased” are examples.2 The importance of disease information garnered from death events was highlighted by John Graunt’s key observations about mortality, which he published in 1662. These would have been impossible without England’s Bills of Mortality.3 John Snow’s contributions to our understanding of cholera, and the very birth of the field of epidemiology, are tied directly to his ingenious use of London’s Bills of Mortality.4 Today, nearly 900 years after the introduction of the death certificate, death data continues to be immensely important in shaping our understanding of the health of a population.
CODING CAUSES OF DEATH
Nearly 5,500 deaths occur daily in the U.S., each spawning a process culminating in a registered death certificate, in which a certifier, typically a physician, has listed the “causes of death”. Nosologists eventually translate the “literals”, unstructured text descriptions of causes of death furnished by certifiers, into codes from the International Classification of Disease (ICD). The manual nature of this process and the volume of data make this a lengthy and laborious process lasting upwards of 8–12 months for more populous states. This process has been facilitated by the National Center for Health Statistics, NCHS, SuperMICAR system, but the ‘coding’ continues to require substantial time and often leaves the local health department manually coding in order to characterize disease patterns quicker than several months after the events. Automated classification of causes from unmodified text literals in death certificates would benefit local jurisdictions significantly in understanding patterns of disease leading to death in their respective communities. In this paper, we describe a series of experiments in which we use the UMLS and statistical text matching algorithms to identify ways of automating the classification of causes of death.
ELECTRONIC DEATH REGISTRATION
Although the death certificate registration process in the U.S. continues to be largely paper-based, a national effort to re-engineer vital records has spurred the development of electronic systems in a number of states. California developed such a system in 2005 and today manages over 99% of its 235,000 annual death certificates electronically. The availability of an automated classification system, engaged perhaps as a web service at the point of recording could have additional benefits in offering an interactive approach to obtaining the causes and improving the quality of death related data.
AUTOMATING CLASSIFICATION
Analysis of unstructured biomedical text and automated classification using key conceptual features is an active area of medical informatics research.5,6,7,8 The automated classification of causes of death from death certificates is a novel use of these techniques. Despite substantial bibliographic research, we were unable to find prior research in this specific application of text classification methods.
For the purposes of our experiments, we conceptualized the classification process as one of matching unstructured input text to existing text descriptions in a coding system. The benefits of this approach are the use of well known and computationally efficient statistical text matching algorithms. However, this approach does not take advantage of semantic relationships that might be available in biomedical terminology systems. Instead, we infer semantic similarity if we match words that are nouns or if non-noun words exceeding a certain length are matched.
TEXT MATCHING APPROACH
Pattern matching is a common approach to string matching and can be classified into edit distance, phonetic analysis and n-gram based techniques.9 Edit distance is useful for short strings where differences are minimal. Phonetic algorithms rely upon the way in which the word is spoken. This approach is highly language specific. The N-gram approach is a better approach for larger strings that can have significant differences. This technique involves generating sub-strings of n-length from the original string, or phrase. The similarity measure between two strings is calculated by calculating the number of n-grams in common. N-grams represent overlapping slices of larger strings. Therefore any errors present in the string will only affect a relative few of its n-gram components thus reducing the impact of misspellings and lexical variations. Biomedical text matching with this technique performs best if one uses at least a tri-gram.10 For our experiments, we used an n-gram size of 4.
METHODS
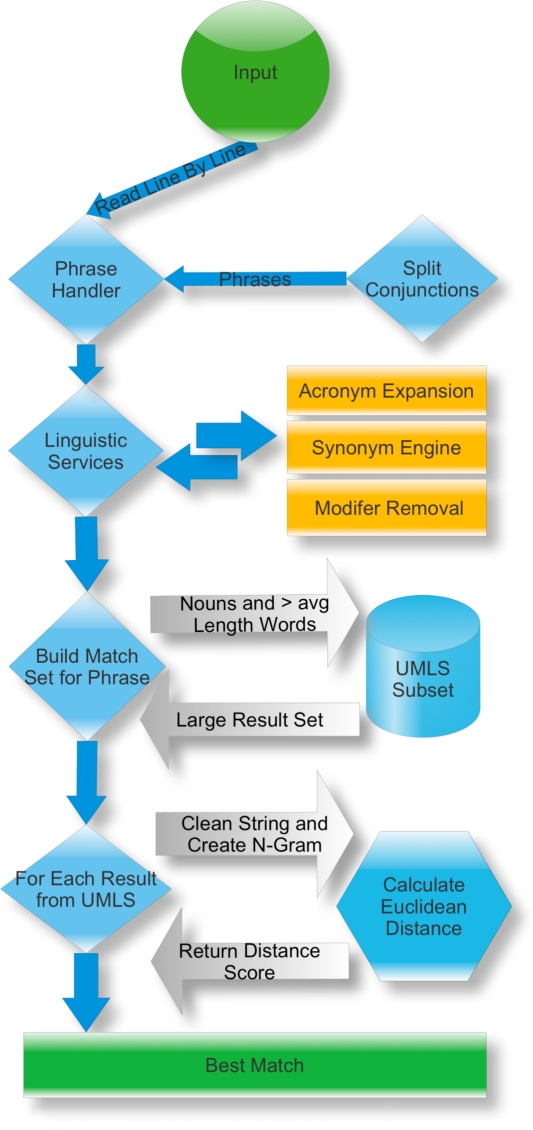
Medical Match Master was built to match unstructured cause of death phrases to concepts and semantic types within the UMLS. The system annotated each input with two types of information, the Concept Unique Identifier, CUI, and a semantic type as assigned by the UMLS. The System utilizes a statistical n-gram approach to resolve matches between input and UMLS phrases. Figure 1 provides a graphic of system features and flow.
Figure 1:
MatchMaster Procedure
Input is fed to the system as a text document, read line by line and processed serially in a pipelined model of execution. The first node, the phrase handler sends each input through the remainder of the pipeline. The next node, linguistic services, employs acronym expansion, a synonym engine and modifier removal to enhance the input while respecting semantic equivalence. The output of the language services node is a set containing the original input string and all of its semantic equivalents. For each phrase in the input set a large match set is constructed by querying the database containing any subset of the UMLS. The next node involves the computation of a distance score for each element in the match set against the input phrase. First, n-grams are generated. During n-gram generation, strings are normalized by converting to lower case and removing spaces, numerics and punctuation. Upon completion of n-gram conversion an optional alphabetization step occurs. This step reduces dependence on word order, and simultaneously reduces dependence on knowledge density within the knowledge source. The resulting n-grams are then compared against one another for similarity using a positional n-gram approach. In this approach, corresponding gram values do not need to occur in the same location index as its counterpart to record a match. Example:
Phrase 1: university
Phrase 2: the university
N-gram 1: uni niv ive ver ers sit ity
N-gram 2: the heu eun uni niv ive ver ers sit ity
The underlined grams represent indices that match. Those not underlined do not register a match. This example shows that a less lenient approach would not have registered any matches. The positional matching only occurs within the number of grams equal to the difference in length between the gram arrays. In this case the algorithm looked up to three indices away.
This method treats each gram value with equal weight. Typical n-gram based techniques utilize a weighted measure for determining the importance of a given n-gram within a document known as tf.idf, or term frequency times the inverse document frequency. The “documents” in this case are sentences. Given their brevity a simple boolean representation of gram significance is used. Either present, 1, or not present, 0. To apply this to the previous example. Consider N-gram 1 and N-gram 2 independently. One Assigns each existing gram a value of one. One then zero pads the shorter string to match the length of the longer. Consider the distance between them to be equal to the square root of the sum of the squares of the difference between them. To reproduce the example from above: +++ has been used to represent the gram values not existent in N-gram 1 that are present in N-gram 2.
+++,0 +++,0 +++,0 uni,1 niv,1 ive,1 ver,1 ers,1 sit,1 ity,1
the,1 heu,1 eun,1 uni,1 niv,1 ive,1 ver,1 ers,1 sit,1 ity,1
For this case the sum of the squares of the distance between the phrases is 3 identifying the square root of 3 as the euclidean distance between these strings. In this example only the case of padding the front end of the string was represented. The additional characters may occur at either the front end or the back end, the system checks both conditions when performing its calculations.
Test set generation
A random set of 1000 anonymized causes of death was obtained from the California Electronic Death Registration System. From each of the 1000 the precipitating cause of death was extracted. The World Health Organization defines the precipitating cause as “the disease or injury which initiated the the train of morbid events leading directly to death.”1 After removing duplicates, the remaining set contained 438 entries. The list was stripped of any nonsensical terms and 300 phrases were randomly selected for use as our test set.
UMLS Preparation
Three subsets of the UMLS were used in the experiments detailed below. Each subset was generated using the Metamorphosys tool provided by the National Library of Medicine, NLM, included with the UMLS download.11 A key benefit to utilization of the UMLS is the ability to create a single vocabulary interface to be re-used across differing subsets. This standardizes the mechanics of querying any UMLS subset. In the case of this system, we focused on two tables within the UMLS schema. The first table MRCONSO contains a unique row for each lexical variant of a given concept. Another table MRXW_ENG contains one row for each unique word in each concept.
The first subset was generated from the UMLS 2009AA release and restricted its contents to only ICD10 sources including ICD10 and ICD10 - PCS. This produced 267,228 rows in MRCONSO and 2,981,700 rows in MRXW_ENG. This subset will be referred to as ICD10 subset.
The second subset was generated from the UMLS 2009AA release and contained all level 0 English terminologies containing more than 1000 concepts and the Systematized Nomenclature of Medicine, SNOMED, a level 9 source. Terminologies with less than 1000 concepts were excluded. Terminologies with specialized intentions other than disease classification were also excluded. This includes all of the genome and anatomical terminologies. This configuration produced 5,672,762 rows in MRCONSO and 19,492,846 rows in MRXNW_ENG. This subset will be referred to as UMLS1 for the remainder of this paper.
The third subset includes every level 0 source plus SNOMED, a level 9 source from the 2009AA release of the UMLS. This configuration produced 4,419,102 rows in MRCONSO and 20,113,519 rows in MRXW_ENG. This subset will be referred to as UMLS2 for the remainder of this paper.
Experiments
Three experiments were conducted, each with a different focus. Experiment 1 was designed to determine the best method for querying the ICD10 subset with SQL queries. This included determining the input phrase features to be used in the query, the table to be queried and, whether or not a wildcard character would be employed. The first goal was the identification of features from the input phrase to be utilized as query terms. This was done in three stages, first we extracted all of the nouns from the input phrases and used them in a wildcard query against all possible strings in the MRCONSO database. Second we repeated this test using all words from the phrase that were greater than the average length of the words within that phrase. Third we repeated this test using a combination approach, either noun or greater than average length terms were used to query the database. Testing demonstrated that a combination approach to be most effective. These results are not surprising given nouns alone may not have the descriptive power to produce a high quality match set.
Experiment 1 had an additional goal of determining which table should be queried to build the best match set. We elected to query the ‘WD’ field of the MRXW_ENG table while maintaining the use of both nouns and lengthier terms from the input phrase. Wildcards were only employed if the word ended in an (‘s) which was stripped and replaced with (%, the MySQL wildcard character).
Experiment 1 also had the goal of determining the impact of alphabetizing the n-grams on match resolution. This goal was accomplished by simply running two versions of each test described above, one version with alphabetized n-grams, the other without. Our testing showed that alphabetization of the n-grams increased match resolution.
Experiment 2 was then conducted to determine the most effective linguistic modifications for improving the performance of the system. This included the addition of an acronym expander, synonym engine, and ambiguous term removal. The output of these modules is a set of semantically interchangeable phrases. These services were employed prior to querying the database. The acronym expander was created to replace acronyms with their expansions. The acronym expansions included in this system were determined by review of output from Experiment 1 testing. The synonym engine was created to build a list of all possible semantically equivalent versions of each input phrase. The terms included in the synonym engine were determined by review of the output from Experiment 1 testing. The ambiguous term service removed unnecessary modifiers from phrases. These included temporal modifiers, severity modifiers, and laterality indicators, also generated from a review of Experiment 1 testing. Testing showed that the addition of these services greatly enhanced match resolution and were therefore included in all future tests.
Experiment 3.0 and 3.1 were designed to test improvement from the use of subsets generated beyond the ICD10 including UMLS1 and UMLS2 while maintaining the same query tactics and linguistic modifications optimized in phases 1 and 2 respectively.
RESULTS
The results are presented in table 1. NumFound represents the number of inputs for which MMM found a match, regardless of match validity. %Found is the proportion of the 300 phrase input set NumFound represents. Following this are 5 rows from left to right which detail the number and type of matches returned by the system. NoMatch indicates the number of phrases in the input set for which both the semantic type and the concept applied by MMM were incorrect. AMB indicates the number of matches that were too ambiguous to classify. In this case it was always possible to relate MMM’s annotation to the input but more information is required to make a conclusion. SemanticMatch indicates the number of phrases for which semantic category was correctly assigned yet the concept is wrong. This was oft the case when dealing with cancers. The system would correctly infer that it was dealing with a neoplastic process and in most cases differentiate between carcinoma, adenocarcinoma, and lymphoma yet frequently indicated the wrong anatomical location or stage of cancer. IncompleteMatch represents the number of cases in which the system correctly identified semantic type yet the concept it returned was of differing specificity than the concept applied. For example, input phrase “cerebrovascular accident” received a concept annotation of “other cerebrovascular diseases”. Since the applied concept annotation is more general than the input it cannot be used to accurately represent the original phrase therefore not deemed a complete match. CompleteMatch represents a valid semantic and concept match. The last two columns provide proportional performance results with respect to their recall(NumFound). %CompleteMatch represents the proportion of completely valid matches the system returned while %AllSem represents the proportion of valid semantic identification.
Noteworthy findings include the improvement in performance between experiments 1 and 2 indicating that the addition of some simplistic linguistic services can provide improvement across the board when utilizing the same UMLS subset. A ∼2% increase in recall was noted from an already high 94.67% to 96.33% and a nearly 5% gain was achieved in the proportion of complete matches. The results also show that by shifting to a more comprehensive subset of the UMLS the proportion of exact matches improves significantly from ∼30% to ∼50%. This is due mainly to a drop in the number of unsuitable matches, indicating that ICD-10 may not have the resolution required to accurately represent causes of death.
| Test | NumFound | %Found | NoMatch | AMB | SemanticMatch | IncompleteMatch | CompleteMatch | %CompleteMatch | %AllSem |
|---|---|---|---|---|---|---|---|---|---|
| Experiment 1.0 | 284 | 94.67 | 69 | 7 | 71 | 58 | 79 | 31.89 | 74.8 |
| Experiment 2.0 | 289 | 96.33 | 64 | 0 | 72 | 50 | 103 | 36.64 | 77.9 |
| Experiment 3.0 | 299 | 99.67 | 34 | 5 | 66 | 40 | 154 | 51.51 | 83.7 |
| Experiment 3.1 | 299 | 99.67 | 42 | 6 | 35 | 61 | 155 | 51.84 | 87 |
DISCUSSION
Our results conclude that with little effort it is possible to identify an exact concept identifier from the UMLS over 50% of the time for cause of death phrases. This alone will greatly speed the reporting process for mortality statistics. We found that the main limiting factor to this approach is the reliance upon knowledge density within the terminologies. We further believe that involving semantic reasoning into the Medical Match Master system will improve its performance.
Acknowledgments
Funded in part by California Department of Public Health
REFERENCES
- 1.WHO . ICD-10: International Statistical Classification of Diseases and Related Health Problems, Tenth Revision. Geneva, Switzerland: 1993. [Google Scholar]
- 2.Cipolla CM. The ‘Bills of Mortality’ of Florence. Population Studies. 1978;32(3):543–8. doi: 10.1080/00324728.1978.10412814. [DOI] [PubMed] [Google Scholar]
- 3.Glass MEO DV, Sutherland I. John Graunt and His Natural and Political Observations [and Discussion] Proceedings of the Royal Society of London. 1963 Dec 10;159(974):2–37. 1963. [PubMed] [Google Scholar]
- 4.Koch T, Denike K. Crediting his critics’ concerns: Remaking John Snow’s map of Broad Street cholera, 1854. Social Science & Medicine. 2009;69(8):1246–51. doi: 10.1016/j.socscimed.2009.07.046. [DOI] [PubMed] [Google Scholar]
- 5.Shu J, Clifford G, Long W, Moody G, Szolovits P, Mark R. An Open-Source, Interactive java-Based System for Rapid Encoding of Significant Event in the ICU using the Unified Medical Language System
- 6.Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB. Frontiers of biomedical text mining: current progress. Brief Bioinform. 2007;8(5):358–75. doi: 10.1093/bib/bbm045. [Author Manuscript] 09/2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barrett N, Weber-Jahnke JH. Applying natural language processing toolkits to electronic health records - an experience report. Stud Health Technol Inform. 2009;143:441–6. [PubMed] [Google Scholar]
- 8.Hazlehurst B, Frost HR, Sittig DF, Stevens VJ. MediClass: A system for Detecting and Classifying Encounter-based Clinical Events in Any Electronic Medical Record. Journal of the American Medical Informatics Association. 2005;12(5) doi: 10.1197/jamia.M1771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Christen P. A Comparison of Personal Name Matching: Techniques and Practical Issues. Canberra: The Australian National University; 2006. [Google Scholar]
- 10.Faber V, Hochberg JG, Kelly PM, Thomas TR, White JM. Concept Extraction a data-mining technique. Los Alamos Science. 1994;(22) [Google Scholar]
- 11.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004 Jan 1;:D267–70. doi: 10.1093/nar/gkh061. 32(Database issue): [DOI] [PMC free article] [PubMed] [Google Scholar]