Abstract
We propose to collect freely available articles from the web to build an evidence-based practice resource collection with up-to-date coverage, and then apply automated classification and key information extraction on the collected articles to provide means for sounder relevance judgments. We implement these features into a dual-interface system that allows users to choose between an active or passive information seeking process depending on the amount of time available.
Background
Evidence-based practice (EBP) is the integration of best research evidence with clinical expertise and patient values1. EBP promotes the synthesis and critical appraisal of healthcare literature to meet the information needs of practitioners, and accelerates the adoption of research findings into practice. It has become commonplace in healthcare in recent years.
The application of EBP can be divided into two stages: 1) evidence gathering and selection, and 2) practice implementation and outcome evaluation. The first stage is crucial because the quality of the evidence gathered directly influences the downstream best practices.
To ensure proper gathering and selection of evidence, most EBP literature suggests an active search process that includes the formulation of clinical questions, the search for evidence and the appraisal of evidence2,3: A clinical question is first formulated using PICO4 (i.e., patient, intervention, comparison and outcome). With this question in mind, keyword searches on EBP resources5 (such as CINAHL, Medline and Embase) are conducted to locate candidate articles. Lastly, the candidate articles are appraised critically, on criteria such as applicability and validity.
Despite the fact that such a proactive process is useful; it is often too difficult and/or time-consuming for the health practitioners due to two reasons:
First of all, EBP resources exist largely in isolation (i.e., do not connect with each other and are only searchable via their own interface) and commonly require subscription. As a result, health practitioners would not only need to perform searches in these resources one by one but also initiate separate searches for the articles which appear to be relevant but are not accessible from the current resource.
Secondly, current search engines are limited in their capabilities for evidence gathering and selection. Publicly accessible generic search engines are able to search through different resource collections and help to find free materials; however, their search results are hard to navigate through unless proper categorization is done to separate different types of resources. In contrast, specialized search engines are designed specifically for medical search with comprehensive medical knowledge and metadata but are often restricted in accessibility and meant to be used exclusively for one particular EBP resource.
Coupled with the fact that most health practitioners have to spend much of their time taking care of patients6 and may not be well-trained in searching, they are often unable to follow this process to keep up with the literature.
An alternative to active search is to postpone the integration of current research practices until later in the healthcare workflow and delegate the selection process to an automated system. For example, some knowledge-based clinical decision support systems link patient records to medical knowledge in knowledge bases to facilitate downstream decision making7. A major problem with such systems is to ensure that the evidence in the knowledge bases always incorporates current research findings8. More recently, meta-search systems such as InfoBot9 present information retrieved from five EBP resources based on the biomedical terms extracted from the patient records. While these systems save the practitioners the trouble of searching through these resources individually, they lack the flexibility to allow the practitioners to customize the search process or explore evidence from other resources.
In summary, for evidence gathering and selection, active search is the recommended practice, but the isolation of EBP resources, as well as the choice between generic and specialized search engine, often complicates the process and makes it less desirable in the interest of time. In contrast, delegating the process to automated systems helps to save time but such systems are challenging to build and often lacking in flexibility and resource coverage.
We propose a system that addresses these limitations. Our system allows healthcare professionals to curate their own sets of relevant articles for EBP, under an organized framework. In this paper we discuss our framework’s three key novel features:
- Feature 1: Harvesting EBP resources by periodic crawling: Our system crawls freely accessible articles from the web to create an EBP resource collection from which professionals can curate materials to their individual liking. This crawling ensures that our collection covers a variety of EBP resources and contains the latest research findings.
- Feature 2: Automated article classification and key information extraction: Automated classification of resources help to filter out irrelevant documents and separate the rest into different categories, assisting a professional to zoom to relevant documents quickly. Key information is additionally extracted to help practitioners more quickly determine the relevance of candidate articles to the formulated clinical questions.
- Feature 3: Dual active/passive user interface: Our system presents two different interfaces to cater for both active and passive search. It allows the practitioners to choose their preferred interaction mode, depending on their goal and time available.
Methods
We will first describe the architecture of our system and then explain the three key features in detail.
System Design
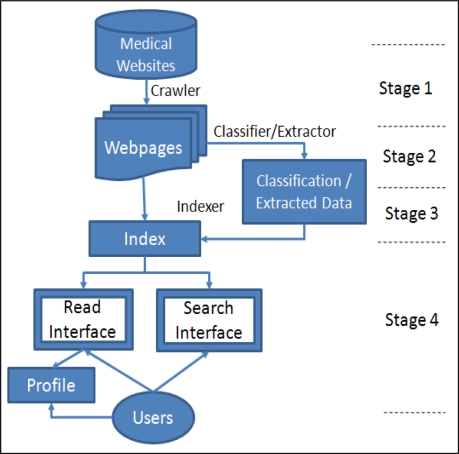
Our system’s architecture (Figure 1) is logically divided into four stages:
Stage 1: We use the Nutch crawler10 to conduct periodic crawls on EBP resources manually selected by experts to obtain a collection of EBP resources.
Stage 2: We then apply machine learned classifiers and extractors to determine the types of the resources and extract various information from them.
Stage 3: We employ Lucene11, a freely-available text search engine library, to index the resources together with the results from classification and extraction.
Stage 4: Practitioners access the information in the index through the Search or Read interface, depending on their information seeking modes.
Figure 1.
System Architecture
Feature 1: Harvesting EBP resources by periodic crawling
To construct the resource collection for our system, we first ask several experts to select a set of websites as the starting points for crawling. Among the selected websites, our system then crawls the contained webpages from the ones that permit crawling. This crawling can be repeated periodically to ensure that the latest documents are ingested.
There are two reasons why we choose to construct our resource collection by crawling: First of all, this method works with all web-accessible materials. As such, our system has no problem harvesting resources of different types or from different websites. This ensures comprehensive coverage of our resource collection. Moreover, periodical crawling addresses the freshness problem, making it trivial to ensure up-to-date information is incorporated in the system.
Feature 2: Automated article classification and key information extraction
While crawling collects webpages from curated sites, not all pages of a site are relevant, primary research. Irrelevant pages, such as table of contents and help pages, are thus filtered from the system. For primary research articles, we determine their types and propagate this information into the downstream user interface, allowing users to choose among different types to view.
As such, we apply supervised learning techniques to classify the pages into three categories: the abstract of a research article, the full text of a research article and any other webpages (to be discarded).
To build the classifiers, we have randomly chosen and annotated 500 webpages from the harvested resources as our training data. We extract token features (e.g., N-grams), webpage features (e.g. URL tokens and content length) and formatting features (e.g., whether a word is in bold/italic) and construct a classification model using Maximum Entropy12. The classification performance of this model (Table 1a) on a 5-fold cross validation13 is indicated by the following standard information extraction metrics:
Precision (P) = TP / (TP + FP),
Recall (R) = TP / (TP + FN),
F1-Measure (F) = 2 * P * R / (P + R),
where TP: true positive, FP: false positive, FN: false negative.
Table 1.
Performance for Webpage Type Classification, Key Sentence Extraction, and Keyword Extraction, Marco-averaged F1 is .98, .48, .81, respectively.
| (a) | P | R | F |
|---|---|---|---|
| Abstract | .95 | .98 | .97 |
| Full text | .94 | .97 | .96 |
| Others | .99 | .98 | .99 |
| (b) | P | R | F |
|---|---|---|---|
| Patient | .70 | .24 | .36 |
| Intervention | .78 | .56 | .65 |
| Result | .90 | .28 | .43 |
| Study Design | .89 | .39 | .54 |
| Research Goal | .92 | .27 | .43 |
| (c) | P | R | F |
|---|---|---|---|
| Sex | .98 | 1 | .99 |
| Condition | .76 | .63 | .69 |
| Race | .92 | .86 | .89 |
| Age | .85 | .78 | .81 |
| Intervention | .74 | .58 | .65 |
| Study Design | .87 | .73 | .80 |
Moreover, through our discussion with professionals, we noticed that other information from the webpage also plays a part in the evidence selection process. Therefore, after classification, we additionally extract the following information from webpages:
Year of publication – Newer publications are preferred, as they present the latest findings. This is extracted using regular expressions.
Time added – The system tracks when resources are added so that it can let users know which resources are new since their last login. This information is obtained directly from crawl data.
URL – Besides serving as a link to the original resource, it also gives the provenance of the resource, which has been shown to be useful in judging its trustworthiness. This information is obtained directly from crawl data.
-
Key sentences – These are sentences in an article that are relevant to well-formed clinical questions. They allow the users to judge the applicability and validity of the article without having to read it in full. The extraction of these sentences is done by a soft classification on the article’s sentences, to determine whether they pertain to five specific types of information: Patient, Intervention, Study Design, Result and Research Goal.
Similar to article type classification, supervised learning based on Maximum Entropy is used for this task. The features employed include token features (e.g., N-grams of the words in the sentence), sentence features (e.g., position the sentence), named entities features (e.g., whether the sentence contains person names), MeSH features (e.g., whether the sentence contains MeSH terms and their categories), and lexica features (e.g., whether the sentence contains words in the manually created wordlist for age.)
The evaluation results (shown in Table 1b) indicate that the sentence extraction is precise, but there is much room for improvement on recall. This performance trend favors our user interface, as the limited screen real estate allows us only to show a limited set of key sentences.
-
Keywords – These are keywords in a key sentence that are relevant to well-formed clinical questions. We follow the same methodology as in sentence extraction, except that the classification is among six categories: Sex, Condition, Race, Age, Intervention and Study Design.
Here, the features employed include token features (e.g., the word itself and its part-of-speech tag), phrase features (e.g., the head noun of the phrase that contains the word), MeSH features (e.g., whether the word is part of a MeSH term) and lexica features (e.g., whether the word appears in the manually created word list for age.)
As shown in Table 1c, our classifier performs well for types that have a fixed and relatively small vocabulary, such as Sex, but can be further improved on categories whose vocabulary is diverse, such as Condition.
A detailed description of the key sentence/keyword extraction is described in a separate work14.
Feature 3: Dual active/passive user interface
Our system keeps users updated with a passive read interface, which recommends relevant articles to them periodically, based on their interests saved in a stored profile. A separate, active searching interface allows the users to pose queries to retrieve relevant articles. The two modes are interlinked, allowing users to change their interaction modes seamlessly.
Read Interface
To make use of the read interface, the healthcare professionals first construct their user profiles. They key in their interests in the form of primary and secondary keywords. The primary keywords represent the topics of interest (usually the names of symptoms or diseases), while the secondary keywords represent the relevant aspects of the topics.
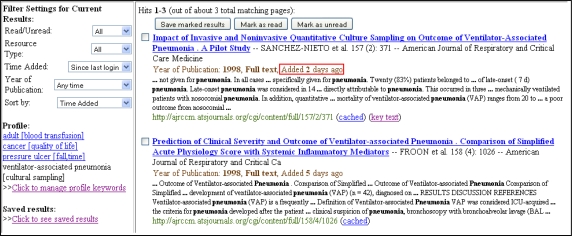
With their interests encoded into the profile, our system automatically presents the latest relevant articles whenever they access the system via the read interface. Figure 2 shows how the read interface highlights recent results that have been added to the system, since the user’s last login. Filtering is also enabled. If a user is only interested in a particular resource type or articles published recently, they can employ filters to customize the results dynamically.
Figure 2.
Read Interface showing latest articles on Ventilator-associated pneumonia
Aside from standard search engine snippet metadata, our system also shows the pertinent extracted information – year of publication, article type, time added, key sentences and keywords (Figure 2 & 3) – assisting users in their relevance assessments.
Figure 3:
Display of the extraction results to assist the users in applicability and validity assessment
Search Interface
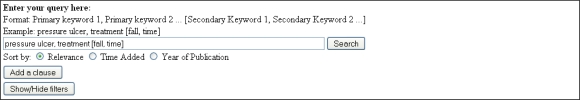
This interface (Figure 4) caters for users who prefer active search and is designed with similar conventions to generic search engines, but with enhanced support for query formulation.
Figure 4.
Query Formulation Tool in Search Interface
Similar to the profile keywords, a query in our system is a combination of primary keywords, which are used to search for articles relevant to a certain topic, and secondary keywords, which are used to filter out articles that are irrelevant to desired aspects of the topic. More complex queries can be constructed by joining multiple queries with Boolean operators. In addition, the users may filter the search results on different facets (e.g., year of publication).
All query-related information, such as the keywords, the time of search and the number of results returned are saved in the search history to help the users to keep track of the searches they have conducted.
Discussion and Future Work
We follow an iterative development methodology in building our system. In each iteration, we meet up with healthcare professionals to study their information seeking behaviors and the problems they have encountered in the evidence gathering and selection process. We then design and implement features to address those problems. When the implementation is finished, we ask them to try out our system and provide feedback on the efficacy of the features. Throughout the whole process, no personal information has been collected, ensuring the protection of the professionals participating.
The healthcare professionals we are working with are the nurses from the Evidence-Based Nursing Unit in National University Hospital. The comments we have received during our informal evaluations with them are mostly positive and encouraging. Among the key features, the classification and extraction feature is most appreciated because it allows them to focus exclusively on the full text articles and see only the latest articles since last login. In some of the evaluation sessions, they also commented that they were able find free full text articles which were not found in other medical databases. This comment is a positive indication that harvesting EBP resources with crawling can be advantageous. Lastly, they expressed interest in the search history function which makes it easier for them to write the search methodology section in their systematic review.
The main challenge we are facing now is that the amount of articles in the collection is still small compared to the known existing EBP resource pool. Among the 94 websites recommended by the nurses, we are only able to crawl 17 of them due to robot exclusion policy. Our current collection contains 3371 full text articles and 16,522 abstracts.
With the current collection, the nurses noted that a more thorough evaluation of the system is possible only if more documents can be indexed so that they could accomplish a realistic, sizeable task – such as a literature review on a concrete topic – with our system. Therefore, we aim to extend our collection and perform a full-fledged user evaluation in future. In this way, our system will become more useful and efficient in the implementation of EBP practices.
Conclusion
We have designed and implemented a literature retrieval system with novel features to facilitate EBP. Initial feedback has been mostly positive and a full-fledged evaluation is planned. As a long-term goal, a study on how the information seeking behaviors differ among different types of healthcare professionals would help to reveal more user- and group-specific features for the system.
References
- 1.Sackett DL, Richardson WS, Rosenberg WMC, Haynes RB. Evidence-based medicine: how to practice and teach EBM. 2nd Ed. London: Churchill-Livingstone; 2000. [Google Scholar]
- 2.Fineout-Overholt E, Melnyk BM, Schultz A. Transforming health care from the inside out: advancing evidence-based practice in the 21st century. J Prof Nurs. 2005;21(6):335–44. doi: 10.1016/j.profnurs.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 3.Brady N, Lewin L. Evidence-based practice in nursing: bridging the gap between research and practice. J Pediatr Health Care. 21(1):53–56. doi: 10.1016/j.pedhc.2006.10.003. [DOI] [PubMed] [Google Scholar]
- 4.Melnyk B, Fineout-Overholt E. Evidence-based practice in nursing and healthcare. Philadelphia: Lippincott; 2005. [Google Scholar]
- 5.Oremann MH. Internet resources for evidence-based practice in nursing. Plastic Surgical Nursing. 2007;27(1):37–9. doi: 10.1097/01.PSN.0000264160.68623.0a. [DOI] [PubMed] [Google Scholar]
- 6.Bond CS. Nurses and computers: an international perspective on how nurses are, and how they would like to be, using ICT in the workplace, and the support they consider that they need. Project Report. London: Florence Nightingale Foundation; 2005. [Google Scholar]
- 7.Bakken S, Currie LM, Lee NJ, Roberts WD, Collins SA, Cimino JJ. Integrating evidence into clinical information systems for nursing decision support. Int J Med Inform. 2008;77(6):413–20. doi: 10.1016/j.ijmedinf.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sim I, Gorman P, Greenes RA, et al. Clinical decision support systems for the practice of evidence-based medicine. J Am Med Inform Assoc. 2001;8(6):527–34. doi: 10.1136/jamia.2001.0080527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Demner-Fushman D, Seckman C, Fisher C, Hauser SE, Clayton J, Thoma GR. A prototype system to support evidence-based practice. AMIA Annu Symp Proc. 2008:151–5. [PMC free article] [PubMed] [Google Scholar]
- 10.Nutch URL: http://lucene.apache.org/nutch/.
- 11.Apache Lucene URL: http://lucene.apache.org/.
- 12.Maxent URL: http://maxent.sourceforge.net/.
- 13.Wikipedia, Cross-validation URL: http://en.wikipedia.org/wiki/Cross-validation.
- 14.Zhao J, Kan MY, Proctor PM, et al. Improving search for evidence-based practice using information extraction. AMIA Annu Symp Proc. 2010 [PMC free article] [PubMed] [Google Scholar]