Abstract
We present a novel user-centric visual analytics system that supports investigation of simulated disease outbreak and the study of decision-making. We developed Epinome as part of our research on decision making in public health and in particular, on the evaluation of information search strategies in public health practice. Epinome is a highly dynamic web-based system that provides a platform to track and study subjects’ decision making and information search strategies, under controlled and repeatable conditions using simulated disease outbreaks. In this paper we focus on the design and implementation of Epinome and present relevant results from field tests we conducted in Utah and Colorado.
Introduction
Effective detection of- and response to infectious disease outbreaks depends both on the ability to capture and efficiently analyze information from disparate sources, and the decisions made by public health officials in response to this information. In the last decade, a multitude of surveillance systems 1,2,3 have been developed to automate data collection, analysis, and generation of statistical alerts or alarms. Time series and geographical information analytics have been the principle modalities employed by these systems for data analysis and presentation8,9. In general, these systems extract and analyze data then present the results as relatively simplistic and predefined visualizations based on what the system developer has deemed important, not on what the user may require. To date, the development focus has been on pushing information to the user, with less attention being paid to the information visualization and data exploration tools needed to optimally support decision-making
In contrast, our focus is on the discourse between the user and the surveillance system, and on the human factors affecting information comprehension and decision-making. Within the context of public health practice, our overall goal is to understand how public health practitioners seek relevant information and improve decision-making in outbreak detection and response. Information search strategies can provide insight into the cognitive biases and heuristics of outbreak investigators (e.g., cognitive tunneling, confirmatory information search). Identification of the presence of such biases will allow for the development of tools that counterbalance these biases and support users in developing alternative scenarios. To support this research, we implemented8 large-scale, high-fidelity simulations of scripted infectious disease outbreaks and developed a novel visual analytics system, named Epinome, to explore the outbreaks and study user interactions. Epinome incorporate information foraging principles10 and empower users to seek relevant information based on their current, and evolving, tasks.
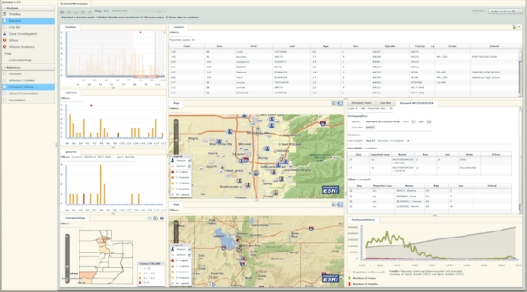
Epinome, shown in Figure 1, is a highly interactive, dynamic and user-centric data visualization workbench designed to facilitate user perception and comprehension of an unfolding outbreak. Epinome supports replay of pre-run, simulated outbreaks with multiple decision points, at which the user can specify which control measures they want to apply (e.g., antibiotics, quarantine, or vaccination). To support information search strategy research, Epinome records users’ interactions, decisions, and management of the workbench environment. In this paper we describe the design, implementation and capabilities of Epinome and present relevant results from field tests we conducted in 2009 with public health practitioners with a wide range of expertise in the states of Utah and Colorado.
Figure 1:
A screen shot of an Epinome display showing various dynamic views of an evolving simulated pertussis outbreak. Both population and individual level data are presented as well as references and simulation guided training tool. The user interactively selects views and arranges them on the display.
Background
Endsley et al.9 defines situational awareness as three separate cognitive levels: perception of the elements in the environment, comprehension of their meaning, and projection of their status in the near future. Endsley has also shown that the overwhelming majority of human cognitive errors occur during the perception stage, with comprehension errors a distance second. Imperfect decision-making, while important, represents only a small fraction of errors. Effective information visualization is a crucial step in the decision-making process. The data presentation should approximate the user’s mental model to simplify the mental transformations required to perceive the data and reduce the cognitive load. The visualization must cater to the user’s current task and empower the user to visualize and correlate the data from multiple views.
User studies are important tools for the study of information search strategies, which can highlight cognitive biases and heuristics of outbreak investigators. A key requirement for conducting psychological experiments is replicability. In the context of this work, replicability means that a simulated outbreak unfolds in the exact same way on every run provided the user made the same policy decisions. To compare the actions taken by different users, such study must ensure all users who followed the same path are presented with the same set of choices at the same times. Finally, the number of possible final outcomes must be restricted for the analysis to have meaningful results.
To ensure replicability and restrict the problem size of the psychological experiments and for training purposes we relied on a scripted, offline, large-scale agent-based simulation of Bordetella pertussis activity in Utah. We developed several outbreak scenarios each with multiple decision points and a limited set of possible policy changes. Multiple runs of each scripted scenario, following all possible paths, were then executed as an offline batch process on a supercomputer. The cognitive study required the number of possible paths to be limited. For this reason the scenarios we developed for the study included two decision points each with three possible policy changes for a total of nine possible paths per scenario and initial conditions. We note that Epinome itself has no limits on the number of decision points or the number of possible policy changes. The data from the various runs along with the scripted scenarios were then stored in a database. The simulation software (SimVis8) and simulation runs were developed and executed by our collaborators at Virginia Bioinformatics Institute at Virginia Tech. The simulation software incorporates a high fidelity representation of social networks and contact patterns, and an advanced Hethcote model of pertussis transmission. The model incorporates vaccination, waning immunity, effects of antibiotic treatment and other control measures (e.g. social distancing), some of which can be variably applied by the user.
System design
The design of Epinome focuses on the human-computer discourse. We employed a user-centric design that emphasizes the role of the system as a dynamic visual analytics support tool as opposed to the more static information boards. Information boards are designed to show predefine summaries or current snapshot of a (possibly changing) data and are thus static in nature. Epinome empowers the user to explore the data from multiple aspects at the same time and support parallel lines of thoughts. Special emphasis is given to simplify user interactions and queries.
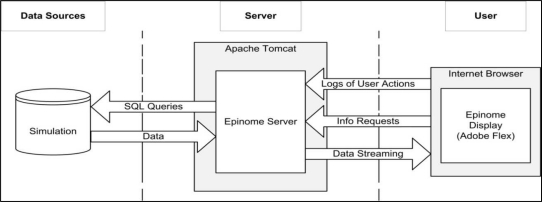
Separation of concerns: The system architecture, shown in Figure 2, is designed to support 1) the need for a dynamic, highly interactive and rapidly responsive user interface, and 2) the need to handle large amounts of heterogeneous data from disparate sources. To balance these needs we developed Epinome as a two-tier web-based system. The presentation layer is implemented as Rich Internet Application (RIA) written in Adobe Flex and runs on virtually any browser under Windows, Mac OS, and Linux. It provides a flexible and responsive user interface that is delivered to the user’s browser at runtime. The backend handles the data access and initial filtering and is implemented as a Java Servlet running under our Apache Tomcat server. During an interactive session, Epinome submits data requests to the server, which in turn fetches data from the database, pre-filters, and streams it back. The system architecture accommodates future extensions to support implementations in public health environments and to enable the use of small-scale dynamic simulation to facilitate ‘what-if’ scenarios.
Figure 2:
System architecture
To examine the effects of various policies and facilitate training we designed scripted scenarios that contained several pre-determined decision points. Decision points include two or more possible policy changes and other user actions, e.g. starting contact tracing, changing vaccination policy for contacts or closing schools. Each scenario was executed multiple times using different random seeds in an offline batch process. For each scenario and initial random seed we ran the simulation through all possible paths. The simulation output for each branch in each path was then saved along with the simulation script in a database.
During playback, when a decision point is reached, the playback pauses and the user is presented with the appropriate options for policy changes and actions based on the scenario description. The user is free to explore the current situation and can use any of the visual analytics capabilities available in Epinome before making a decision. Once the user decides on a course of action, the system fetches the simulation output associated with the selected branch and resumes the playback from the backend server. From the user’s point of view there is no difference between a simulation playback and a simulation that runs online. In fact the user need not be aware that the outbreak simulation is a playback of pre-computed simulations.
User-centered design
Our goal in the design of the user interface was to provide a rich and dynamic environment in which a user can explore, compare and analyze geo-temporal data, and consult additional internal and external reference knowledge. In that respect, the environment encompasses capabilities and methods of both task environment and information environment10. In a task environment the user motivation is based on a goal or a task; the detection of an outbreak in our case. An information environment, on the other hand, permits the user to engage their tasks more adaptively, to seek additional knowledge, and to become better informed before taking any specific action.
To achieve a friendly blend between task and information environments, the main interface feature a workspace in which the user can place various tools, such as line lists, epicurves and maps, to present different views of the data. As the user’s focus of attention and tasks change, the user can drag additional tools into the workspace, interactively resize and rearrange them within the workspace area, organize them in folders, or remove tools that are not needed any more. This allows users to adapt and transform the display to support the nonlinear workflow of hypothesis-driven outbreak investigation.
Within Epinome the user can have multiple workspaces, each with its own collection of tools centering on different workflows. For example, the user can play multiple simulation scenarios in parallel, each within its own workspace. This provides the ability to dynamically shift focus between parallel lines of thought. The role of a workspace is to share data between the various active tools and coordinate the interactions between them. In addition to the regular workspace Epinome contains other types of workspaces for controlling playback of outbreak simulations and for studying user information seeking strategies.
Available tools
Epinome’s tools are based on common epidemiologic methods of describing public health data. Examples of currently available tools include the following: epicurve, line lists, case investigation forms, demographics information such as schools and the students that attend them, contact tracing reports and interactive point and choropleth maps using an ESRI map server. Other tools provide static references, diagrams and charts relevant to the disease of interest. The user can use multiple instances of the same tool, for example, multiple maps each focusing on a different geographical area or different data type.
Interaction logging
During monitored sessions, the user’s interactions are sent to the remote server and are stored in the database. The logged events can then be explored and analyzed offline by the research team. Examples of logged events include creation, and deletion of views, controlling simulation runs and applying filters to various views.
Results
We conducted a field test to study search strategies in public health practice. The study encompassed 27 public health workers as described in Table 1. Participants were selected from staff directories of state and local health departments in Colorado and Utah. Those who indicated that they had outbreak investigation experience were eligible to participate.
Table 1:
Field test participants
| N=27 | |
|---|---|
| Age group | |
| 18–29 | 3 |
| 30–39 | 8 |
| 40–49 | 7 |
| 50–59 | 6 |
| 60+ | 3 |
| Male | 14 |
| Years of experience in communicable diseases | |
| <2 | 2 |
| 2 – <4 | 4 |
| 4 – <10 | 8 |
| 10+ | 13 |
| Job position | |
| Director or executive director | 9 |
| Manager | 9 |
| Front line | 9 |
Participants investigated multiple simulated outbreaks and took advantage of all the tools provided by Epinome. They used epicurves, line lists and both point and choropleth maps to enhance overall situational awareness. Case investigation forms, school reported case forms and school summaries were used for drill down and contact tracing to investigate potential outbreaks in suspected schools. Participants successfully employed filtering capabilities to focus on populations of interest. As the focus of their investigation shifted, users dynamically created new views (tools) and rearrange them on the screen to maximize the amount of relevant information displayed on the screen.
The focus of the cognitive field study was on information search strategies; it was not a formal usability test. We did, however, ask several ease of use and utility questions as part of the study. Participants rated Epinome highly for its ease of use and ability to provide insight. They also expressed a desire to use Epinome in the workplace, as shown in Table 2. The full description of the field tests, analysis and evaluation of information search strategies will be presented in a later paper.
Table 2:
Comments from field test participants about the usability and usefulness of Epinome.
| Disagree Strongly | 2 | 3 | 4 | 5 | 6 | Agree Strongly | Response Average | |
|---|---|---|---|---|---|---|---|---|
| 1 | 7 | |||||||
| Provided me with new insights | 0 | 0 | 0 | 1 | 4 | 6 | 14 | 6.32 |
| Valuable decision support tool | 0 | 0 | 0 | 0 | 1 | 10 | 14 | 6.52 |
| Highly usable | 0 | 0 | 0 | 1 | 2 | 7 | 15 | 6.44 |
| Would help me do my job more efficiently | 0 | 0 | 1 | 2 | 1 | 6 | 15 | 6.28 |
| I would benefit from having a tool like Epinome to support my work | 0 | 0 | 0 | 1 | 2 | 6 | 16 | 6.48 |
Conclusion and Future Directions
We present a novel user-centric visual analytics system that supports investigation of simulated disease outbreaks and the study of decision making in public health. We describe the requirements, overall design and implementation of the system. The system performed reliably running remotely over the web during field tests in Utah and Colorado. Participants in the field study reported overwhelming enthusiasm for the capabilities and usability of the system.
The lessons learned from the field study are being incorporated into ongoing efforts to refine and extend the set of tools in Epinome both in number and in scope. We are now working with the Utah Department of Health to extend Epinome to handle NEDSS data and to implement Epinome as a practical application for public health practitioners.
References
- 1.Lombardo JS, Burkom H, Pavlin J. ESSENCE II and the Framework for Evaluating Syndromic Surveillance Systems. Morbidity and Mortality Weekly Report. 2004;53:159–165. [PubMed] [Google Scholar]
- 2.Gesteland PH, Samore MH, Pavia AT, Srivastava R, Korgenski K, Gerber K, Daly JA, Mundorff MB, Rolfs RT, James BC, Byington CL. Informing the front line about common respiratory viral epidemics. AMIA Annu Symp Proc. 2007 Oct 11;:274–8. [PMC free article] [PubMed] [Google Scholar]
- 3.Wagner MM, et al. Syndrome and Outbreak Detection using Chief-Complaint Datt: Experience of the Real-Time Outbreak and Disease Surveillance Project. Morbidity and Mortality Weekly Report. 53:28–31. 3004. [PubMed] [Google Scholar]
- 4.Jinangping Shuai, Pete Buck, Paul Sockett, Jeff Aramini, Pollari Frank. A GIS-driven integrated real-time surveillance pilot system for national West Nile virus dead bird surveillance in Canada. Inter J of Health Geographics. 2006 doi: 10.1186/1476-072X-5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng D, Chen H, Tseng L, Larson C, Eidson M, Gotham I, Lynch C, Ascher M. West Nile Virus and Botulism Portal: A Case Study in Infectious Disease Informatics, in Intelligence and Security Informatics, Proceedings of ISI-2004, Lecture Notes in Computer Science. In: Chen H, Moore R, Zeng D, Leavitt J, editors. Vol. 3073. Springer; 2004. pp. 28–41. [Google Scholar]
- 6.EDEN (Emerging Diseases in changing European eNvironment) http://www.eden-fp6project.net/
- 7.Markus Reinhardt, Johannes Elias, Albert Jürgen, 1, Frosch Matthias, Harmsen Dag, Vogel UlThankrich. EpiScanGIS: an online geographic surveillance system for meningococcal disease. [DOI] [PMC free article] [PubMed]
- 8.Barrett C, Eubank S, Marathe M. The New Paradigm. Springer Verlag; 2005. Modeling and Simulation of Large Biological, Information and Socio-Technical Systems: An Interaction Based Approach, in Interactive Computation. [Google Scholar]
- 9.Endsley Mica R, Bolté Betty, Jones Debra G. Designing For Situational Awareness, An Approach to User-Centered Design. Taylor & Francis; 2003. [Google Scholar]
- 10.Peter Pirolli. Information Foraging Theory: Adaptive Interaction with Information. Oxford University Press; 2007. [Google Scholar]