Abstract
Mild Cognitive Impairment (MCI) is thought to be a precursor to the development of early Alzheimer’s disease (AD). For early diagnosis of AD, the development of a model that is able to predict the conversion of amnestic MCI to AD is challenging. Using automatic whole-brain MRI analysis techniques and pattern classification methods, we developed a model to differentiate AD from healthy controls (HC), and then applied it to the prediction of MCI conversion to AD. Classification was performed using support vector machines (SVMs) together with a SVM-based feature selection method, which selected a set of most discriminating predictors for optimizing prediction accuracy. We obtained 90.5% cross-validation accuracy for classifying AD and HC, and 72.3% accuracy for predicting MCI conversion to AD. These analyses suggest that a classifier trained to separate HC vs. AD has substantial potential for predicting MCI conversion to AD.
INTRODUCTION
Alzheimer’s disease (AD) is the most common cause of dementia. Living longer is putting more people at risk for AD. Deaths from AD have increased significantly, in contrast to deaths from other diseases such as many types of cancers which have dropped1.
Despite incidence rates doubling every 5 years after the age of 65, no treatment currently is available to slow or stop the deterioration of brain cells in AD1. Early diagnosis could facilitate disease-modifying treatments for AD to help delay progression. Therefore, it would be of great potential value to develop better diagnostic tools that can recognize AD at early symptomatic and especially pre-symptomatic stages. To this end, amnestic mild cognitive impairment (MCI) has been defined as a prodromal stage intermediate between healthy controls (HC) who are cognitively normal and individuals with a clinical diagnosis of probable AD2–3. MCI is generally thought to be a precursor to the development of early AD, because patients with MCI have an increased probability of developing AD with a conversion rate of approximately 15% per year2–3. As a result, MCI has received a lot of attention in a wide variety of clinical and research studies. For early diagnosis of AD, it is a challenging problem to predict those who are mostly likely to convert from MCI to probable AD. As MCI does not fulfill current criteria for AD, standard clinical and psychometric assessments currently used for diagnostic criteria for AD are insufficient for this specific goal. Structural magnetic resonance imaging (MRI) has increasingly been used in research contexts to support the clinical identification of AD, or progression to AD, at an earlier stage than standard neurological diagnosis. Regional brain atrophy often begins long before AD is clinically detectable. Moreover, automatic or semi-automatic techniques for analyzing high-resolution structural MRI data have now been developed, such as voxel-based morphometry (VBM) (http://www.fil.ion.ucl.ac.uk/spm/) and brain segmentation and parcellation approaches such as FreeSurfer (http://surfer.nmr.mgh.harvard.edu/).
There have been a number of reports of classification approaches attempting to separate AD and HC or to discriminate MCI from HC using whole-brain MRI analyses or a pre-defined subset of brain regions such as the hippocampus4–11. Most prior studies have been limited by small samples or they did not predict which subjects with MCI would progress to a diagnosis of AD4–9. In addition, some prior studies10–11 investigated prediction of MCI conversion to AD by learning the classifier directly from two MCI subgroups: MCI-Stable (MCI-S) and MCI-Converter (MCI-C). The MCI-C group includes individuals who were diagnosed with MCI at baseline and converted from MCI to probable AD after baseline. The reported highest accuracy is 94.5% for classifying AD vs HC6 and 81.5% for MCI-C vs MCI-S11.
The goal of the present study is to predict MCI conversion to probable AD. Unlike many of prior studies, we train a classifier using data from AD and HC, and then apply it to predicting MCI conversion to AD in an independent set of MCI individuals from the same study assessed using the same methods. The classification accuracy rate was calculated at three different longitudinal time points. Furthermore, we combined imaging features extracted from two different whole-brain analysis techniques (VBM and FreeSurfer) and performed feature selection to identify variables with predictive power, resulting in an improved accuracy for classification. We analyzed data from a large cohort of extensively characterized and imaged subjects from the Alzheimer’s Disease Neuroimaging Initiative (ADNI).
MATERIALS AND METHODS
Subjects
All subjects used in this study are participants of Alzheimer’s Disease Neuroimaging Initiative (ADNI) (http://www.adni-info.org). The ADNI was launched in 2003 to help researchers and clinicians develop new treatments for MCI and early AD, monitor their effectiveness, and lessen the time and cost of clinical trials. Neuroimaging and biological markers were used to achieve the goal of the ADNI study. This 5-year multi-site longitudinal study was started by the National Institute on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), the Food and Drug Administration (FDA), private pharmaceutical companies, and non-profit organizations. The ADNI participants consist of AD, MCI, and elderly HC. They were aged 55–90 years and recruited from 59 sites across the U.S. and Canada. We divided the ADNI cohort into four groups by baseline diagnosis and the MCI to probable AD conversion status using follow-up diagnosis up to 3 years: HC, MCI stable (MCI-S), MCI converter (MCI-C), and AD12. Written informed consent was obtained from all participants and the study was conducted with prior approval from Institutional Review Boards at all sites. For the clinical diagnosis of AD, National Institute of Neurological and Communicative Disorders and Stroke and the Alzheimer’s Disease and Related Disorders Association (NINCDS-ADRDA) criteria, Mini Mental State Examination (MMSE) scores, Clinical Dementia Rating (CDR), and other cognitive assessments were used. Demographic information, APOE genotype, neuropsychological test scores, and diagnosis were downloaded from the ADNI clinical data repository (http://www.loni.ucla.edu/ADNI/).
Image processing and feature extraction
3D T1-weighted brain MRI scans were acquired using a sagittal 3D MP-RAGE sequence following the ADNI MRI protocol. Brain-wide target imaging features from all MRI scans of ADNI participants were processed and extracted using two fully automatic methods as detailed in previous studies12–13. SPM5 (http://www.fil.ion.ucl.ac.uk/spm/) was used for VBM analysis to create an unmodulated and normalized gray matter (GM) density map in the Montreal Neurological Institute (MNI) space and then to extract a single mean GM density value for 86 regions of interest (ROIs) in MNI space. In addition, FreeSurfer V4, an automatic brain segmentation and cortical parcellation tool, was used to label cortical and subcortical tissue classes using an atlas-based Bayesian segmentation procedure and to extract volumetric and cortical thickness values for 56 ROIs in addition to total intracranial volume (ICV).
Pattern classification methods
The support vector machine (SVM) is a classification method for supervised learning. As a powerful and popular multivariate classification algorithm, SVM has been widely used in a variety of classification tasks and produced superior empirical results14.
SVM seeks a linear decision surface (optimal separating hyperplane), which separates two classes of training samples and maximizes the distance (margin) between the decision boundary and the closest samples (support vectors) in each class.
If an effective linear decision surface cannot be identified, the data can be nonlinearly mapped into a higher dimensional space (feature space) in order to gain additional discriminative power. By choosing an appropriate mapping, the data may become linearly separable or mostly linearly separable in the high-dimensional feature space. To this end, various kernel functions can be used to transform the data.
We used radial basis function kernels (RBF). These kernels have two parameters: a cost parameter (C) and a parameter (γ). To determine the optimal values C and γ of the SVM, we used a grid-search. Using a 10-fold cross-validation procedure, we performed classification with each set of parameters (γ, C) varying along a grid ranging from C=20 … 210 and γ=2−10 … 20 to estimate the prediction accuracy. The optimal γ and C were then used to create the SVM model14.
Continuous numeric predictor variables were normalized to have zero mean and unit variance by subtracting the sample mean and dividing by the sample standard deviation. This normalization helped ensure these variables received equal consideration by the modeling and feature selection processes since prediction with large numeric range generally dominate those with smaller range. Note that the mean and sample standard deviation values used to normalize the training data were also used to normalize the testing data to ensure consistent scaling.
Due to the large number of predictor variables, it was important to identify a subset of predictor variables which maximizes the performance of the predictor and results in an improved classification rate. The feature selection was done using an SVM-based criterion, SVM-RFE (SVM-Recursive Feature Elimination)15. SVM-RFE is a simple, efficient and established algorithm that performs feature selection via a sequential backward elimination procedure. The SVM-RFE algorithm returns a ranking of all the features. SVM-RFE ranks features based on the weights of a linear SVM.
RESULTS
In this study, we used all the ADNI participants whose MRI scans passed both VBM and FreeSurfer processing pipelines. Each subject has 142 imaging ROI variables (56 from FreeSurfer and 86 from VBM). All the imaging ROI features were adjusted for the baseline age, gender, education, handedness, and intracranial volume (ICV) using the regression weights derived from the HC participants. In addition to 142 imaging features, family history for dementia and the status of APOE (presence or absence of APOE ɛ4 (or ɛ2) allele) were included in our models. Table 1 displays the number of subjects used in this study for each diagnosis group. We used the diagnosis results at baseline, 12 month (1 year), 24 month (2 year), and at the last visit as of January 23, 2010 (3 year).
Table 1.
Number of subjects used in this study for each diagnosis group
| HC | MCI | AD | ||
|---|---|---|---|---|
| MCI-C | MCI-S | |||
| Baseline | 226 | 389 | 182 | |
| 1 year | 62 | 278 | ||
| 2 year | 110 | 157 | ||
| current | 150 | 205 | ||
Classification between AD and HC
First, SVM was applied to the ADNI imaging ROI data to train the classifier for distinguishing AD from HC at baseline. All analyses were performed using 7-fold cross-validation to avoid over-training of classifiers. In each run, the accuracy, sensitivity, and specificity were calculated as a function of the number of selected features in descending order from the ranked feature list. The 7-fold cross-validation process was repeated 3 times using randomly divided data sets in each iteration. The classification accuracy is shown in Table 2. As expected, the best classification was obtained by using both the FreeSurfer and VBM features, giving a correct classification rate of 90.4%, a sensitivity of 85.0%, and a specificity of 94.8%.
Table 2.
Classification results of AD from HC
| Date set | Accuracy | Sensitivity | Specificity |
|---|---|---|---|
| FreeSurfer | 89.7 | 85.3 | 93.2 |
| VBM | 84.4 | 77.5 | 89.5 |
| FreeSurfer + VBM | 90.5 | 85.0 | 94.8 |
Prediction of MCI conversion to probable AD based on the AD-HC classification model
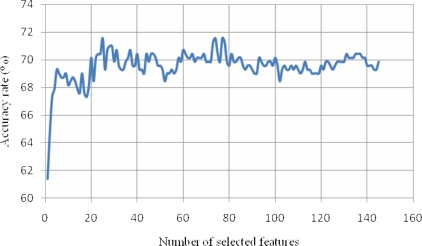
After a classification model was constructed from the AD and HC groups, the same model was applied to the classification of MCI-C versus MCI-S. The classification accuracy rate was calculated at three different longitudinal time points: 1, 2 years after baseline, and January 23, 2010 (3 year). Since the testing data (MCI-C and MCI-S) is different from the training data (HC and AD), no resampling cross-validation process was necessary. The classification results are shown in Table 3. The MCI conversion to probable AD until 1 year after baseline can be predicted using the VBM ROIs with an accuracy rate of 65.0% which is better than the other data sets (FreeSurfer and FreeSurfer+VBM). The MCI conversion to probable Alzheimer’s disease 2 or 3 years later was best predicted using all ROIs from both FreeSurfer and VBM yielding accuracy rates of 72.3% and 71.6%, respectively. A ranked feature list was determined using SVM-RFE. Figure 1 shows the prediction accuracy as a function of the number of selected features in descending order from the ranked features. The data points in Fig. 1 were calculated using all the features of both FreeSurfer and VBM and most recent diagnosis (01/23/2010). The best prediction of MCI conversion to probable AD was obtained for a number of features between 24 and 26. The most important feature identified by SVM-RFE is left entorhinal cortical thickness. Left entorhinal cortical thickness can predict the MCI conversion to probable AD at accuracy of 61.4%. The 25 best features consisted of 7 FreeSurfer ROIs, 16 VBM ROIs, and the status of APOE ɛ4 and ɛ2 genotypes. We obtained a prediction accuracy of 69.3% using the first 5 of these features: thickness of left entorhinal cortex, right hippocampal volume, APOE ɛ4 status, mean temporal lobe thickness (of inferior, middle, and superior temporal gyri), and gray matter density of left hippocampus.
Table 3.
Classification results of MCI-C from MCI-S based on the classifier of AD from HC
| Data set | FreeSurfer | VBM | FreeSurfer + VBM | ||||||
|---|---|---|---|---|---|---|---|---|---|
| 1 year | 2 year | 3 year | 1 year | 2 year | 3 year | 1 year | 2 year | 3 year | |
| Accuracy | 60.3 | 69.7 | 71.5 | 65.0 | 67.8 | 70.7 | 63.8 | 72.3 | 71.6 |
| Sensitivity | 80.6 | 75.5 | 78.0 | 75.8 | 65.5 | 72.0 | 75.8 | 78.2 | 75.3 |
| Specificity | 55.8 | 65.5 | 66.8 | 62.6 | 69.4 | 69.8 | 61.2 | 68.2 | 68.8 |
Figure 1.
Classification of MCI-C from MCI-S based on the classification model of AD vs. HC. The prediction accuracy rate is shown as a function of the number of selected features in descending order from a ranked feature list.
DISCUSSION AND CONCLUSION
Using automatic whole-brain MRI analysis techniques and pattern classification methods, we predicted conversion from mild cognitive impairment to probable AD based on a classification model learned from AD and HC data. In this study, we used MRI scans from 797 participants in the ADNI cohort. We obtained better results when we combined cortical thickness, volume, and gray matter density measures determined from two automatic whole-brain analysis techniques. In addition, the best prediction accuracies were obtained using a subset of features identified and ranked by a feature reduction algorithm. The most important three features are left entorhinal cortical thickness, right hippocampal volume, and APOE ɛ4 status1. It has been shown that regional brain atrophy occurs initially and most severely in the entorhinal cortex and hippocampus before spreading throughout the neocortex16. APOE ɛ4 is a well established genetic risk factor for Alzheimer’s disease. We conclude that a classifier trained to classify AD from HC has substantial potential for predicting MCI conversion to AD. These results encourage further investigation of algorithms for determining those at greatest risk for disease progression. This is important for identifying those patients who might benefit most from a clinical trial or as a stratification approach within clinical trials.
Acknowledgments
Data collection and sharing was funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI; PI: Michael Weiner; NIH grant U01 AG024904). ADNI is funded by the National Initiative on Aging (NIA), the National Institute of Biomedical Imaging and Bioengineering (NIBIB), and through generous contribution from the following: Pfizer Inc., Wyeth Research, Bristol-Myers Squibb, Eli Lilly and Company, GlaxoSmithKline, Merck & Co. Inc., AstraZeneca AB, Novartis Pharmaceuticals Corporation, the Alzheimer’s Association, Eisai Global Clinical Development, Elan Corporation plc, Forest Laboratories, and the Institute for the Study of Aging, with participants by the U.S. Food and Drug Administration. Industry partnerships are coordinated through the Foundation for the National Institute of Health. The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Disease Cooperative Study at the University of California, San Diego. ADNI data are disseminated by the Laboratory of Neuro Imaging at the University of California, Los Angeles. This work also was funded by grant U24AG021886 from National Cell Repository for Alzheimer’s Disease.
Data analysis was supported in part by NLM T15LM07117, NIBIB R03 EB008674, NIA R01 AG19771, NCI R01 CA101318 and U54 EB005149 from the NIH, Foundation for the NIH, and grant #87884 from the Indiana Economic Development Corporation (IEDC).
Abbreviations:
- MCI
mild cognitive impairment
- AD
Alzheimer’s disease
- HC
healthy control
- ADNI
Alzheimer’s Disease Neuroimaging Initiative
- MRI
magnetic resonance imaging
- VBM
voxel-based morphometry
- SPM
statistical parametric mapping
- ROI
region of interest
- MP-RAGE
magnetization prepared rapid acquisition gradient echo
- APOE
apolipoprotein E
- SVM
support vector machine
- GM
gray matter
- SVM-RFE
support vector machine-recursive feature elimination
- MNI
Montreal Neurological Institute
REFERENCES
- 1.Alzheimer’s Disease Facts and Figures. http://www.alz.org
- 2.Peterson RC, Smith GE, Waring SC, Ivnik RJ, Tangalos EG, Kokmen E. Mild cognitive impairment: clinical characterization and outcome. Arch Neurol. 1999;56:303–8. doi: 10.1001/archneur.56.3.303. [DOI] [PubMed] [Google Scholar]
- 3.Petersen RC, Roberts RO, et al. Mild Cognitive Impairment: Ten Years Later. Arch Neurol. 2009;66:1447–55. doi: 10.1001/archneurol.2009.266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Vemuri P, Gunter JL, Senjem M, et al. Alzheimer’s disease diagnosis in individual subjects using structural MR images: Validation studies. NeuroImage. 2008;39:1186–97. doi: 10.1016/j.neuroimage.2007.09.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kloppel S, Stonnington CM, et al. Automatic classification of MR scans in Alzheimer’s disease. Brain. 2008;131:681–89. doi: 10.1093/brain/awm319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Magnin B, Mesrob L, Kinkingnehun S, et al. support vector machine-based classification of Alzheimer’s disease from whole-brain anatomical MRI. Neuroradiol. 2009;51:73–83. doi: 10.1007/s00234-008-0463-x. [DOI] [PubMed] [Google Scholar]
- 7.Fan Y, Batmanghelich N, et al. Spatial patterns of brain atrophy in MCI patients, identified via high-dimensional pattern classification, predict subsequent cognitive decline. NeuroImage. 2008;39:1731–45. doi: 10.1016/j.neuroimage.2007.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gerardin E, Chetelat G, et al. Multidimenstional classification of hippocampal shape features discriminates Alzheimer’s disease and mild cognitive impairment from normal aging. Neuroimage. 2009;47:1476–86. doi: 10.1016/j.neuroimage.2009.05.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chupin M, Gerardin E, Cuingnet R, et al. Fully automatic hippocampus segmentation and classification in Alzheimer’s disease and mild congnitive impairment applied on data from ADNI. Hippocampus. 2009;19:579–87. doi: 10.1002/hipo.20626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Querbes O, Aubry F, et al. Early diagnosis of Alzheimer’s disease using cortical thickness: impact of cognitive reserve. Brain. 2009;132:2036–47. doi: 10.1093/brain/awp105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Misra C, Fan Y, Davatzikos C. Baseline and longitudinal patterns of brain atrophy in MCI patients, and their use in prediction of short-term conversion to AD: Results from ADNI. Neuroimage. 2009;44:1415–22. doi: 10.1016/j.neuroimage.2008.10.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Risacher SL, Saykin AJ, West JD, Shen L, Firpi HA, McDonald BC. Baseline MRI predictors of conversion from MCI to probable AD in the ADNI cohort. Curr Alzheimer Res. 2009;6:347–361. doi: 10.2174/156720509788929273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shen L, Kim S, Risacher SL, Nho K, et al. Whole Genome Association Study of Brain-Wide Imaging Phenotypes for Identifying Quantitative Trait Loci in MCI and AD: A Studt of the ADNI Cohort. NeuroImage. 2010;2010;01:042. doi: 10.1016/j.neuroimage.2010.01.042. doi:10.1016/j.neuroimage. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Vapnick V. The nature of statistical learning theory. New York: Springer Verlag; 1995. [Google Scholar]
- 15.Guyon I, et al. Gene selection for cancer classification using support vector machines. Mach Learn. 2002;46:389–422. [Google Scholar]
- 16.Scahill RI, Schott JM, et al. Mapping the evolution of regional atrophy in Alzheimer’s disease: Unbiased analysis of fluid-registered serial MRI. PNAS. 2002;99:4703–07. doi: 10.1073/pnas.052587399. [DOI] [PMC free article] [PubMed] [Google Scholar]