Abstract
The Internet has become a common source for consumers to seek health information across a wide range of topics including searching for clinical trials. However, not much is known about what consumers search for in relation to clinical trials and how they formulate their search queries. In this study, we use log file data from TrialX.com, a consumer-centric website that provides clinical trial information to ascertain patterns in consumer queries. We analyzed semantic patterns in the queries by mapping query keywords to the UMLS Semantic Types and performed a manual evaluation of user paths. We found that the queries can be grouped into combinations of information needs related to condition, location and treatment. The results also suggested that the consumers using longer search queries with multiple Semantic Types are more likely to take action to participate in clinical trials. The study provides early insights that can be used to inform changes in website content and information display to improve clinical trials information seeking.
Introduction
In the last decade, the Internet has grown tremendously and changed the way consumers obtain health information or engage in health discussions1. As per Pew, 80% of online Americans search for health information on the Internet2 and an overwhelming majority of patients (75%) feel reassured or empowered with the ability to search and satisfy their information needs. The health related searches range across several topics including medical conditions, medications, providers or clinical trials. Various websites (WebMD.com, MedlinePlus.gov), health discussion forums (MedHelp.org) and search engines (Google, Yahoo) have emerged as frequently used resources to obtain general health information. ClinicalTrials.gov is one of the primary sources of information on clinical trials that receives hundreds of thousands of visitors every month3.
Previous research has elucidated general trends related to online health information seeking2, theories on how patients formulate their search queries4,5 what terms they use to search for health information6, how they navigate a clinical trial listing site7 or the kind of queries that remain unsatisfied8. However, not much is known about what patients actually search for in relation to clinical trials or what terms they use to phrase their queries. There are several reasons to ascertain the information needs of health consumers as they seek information about clinical trials. One, determining what terms consumers use to find clinical trials would help identify the information gaps in what they are looking for and what information is available and the language that information is presented in. It has been shown that consumers use short length queries (sometime single, two word phrases) to find information, which may lead to many non-specific results, or use terms without fully realizing that those terms may not be appropriate for their information needs6. Additionally, even though, websites such as ClinicalTrials.gov are routinely used by patients to find clinical trials, studies have reported that they contain information that is technical and written at a reading grade level that is much higher than the average consumer, further illustrating the gap between what patients may be looking for and what is available9. Two, understanding information needs of the end system users is critical in designing useful information systems. This is particularly important for online information resources, because patients use heuristics to quickly ascertain if a website serves their information needs. One of the factors involved in making this decision is the perception of relevance regarding the content of the website10. Three, tailored health communication has been effective in promoting health behavior change11 and it is likely that tailoring clinical trial information based on the consumers knowledge of clinical trials and health condition may play a role in motivating them to participate in clinical trials.
Besides the above reasons, in general, improvements in consumer information seeking for clinical trials information has potential for significant overall impact – that is speeding the development of new treatments. It is estimated that 80% of clinical trials are delayed and most often these delays are due to lack of finding eligible participants12. Only 3% of cancer patients enroll in clinical trials despite 76% of them wanting to enroll in them, if they had the right information13. Improving consumers’ access to clinical trials information in a manner that is suited to their information seeking behavior can be critical in increasing the pool of participants who wish to engage in clinical trials and thus reduce some of the current bottlenecks.
In this paper we report on our analysis to understand information seeking behavior of health consumers in relation to clinical trials. The goal of this research is to expand the current knowledge base of clinical trials information needs among health consumers by performing an in-depth analysis of keyword based queries. To achieve this goal, we have examined the web server logs from an online clinical trial information website. We studied various characteristics such as the length of the query, frequency of keywords, Semantic Type patterns of the keywords to develop a comprehensive information seeking model of clinical trials.
Dataset and Resources
TrialX
TrialX (http://www.TrialX.com) is a website (managed by Applied Informatics Inc.) that provides online services for patients and clinical trial investigators to find and connect with each other. Patients can search for clinical trials by manually entering their health information (condition, age, gender, medication) or trial related parameters (phase, treatment, site location). The patients can also use their Personal Health Record (from Microsoft HealthVault, HealthVault.com or Google Health, Google.com/health) to automatically find the matching clinical trials. TrialX lists all active clinical trials available on ClinicalTrials.gov, CenterWatch.com and trials entered manually by investigators using the service. TrialX pages are indexed across web search engines (Google, Yahoo, Bing, AOL) and the website receives several thousand unique visitors every month, with a majority arriving on the website via keywords typed on search engines. The TrialX website is hosted on an Apache 2 web server and all the web requests are logged. The logs contain a referrer field that provides information about the query typed on the search engine before the visitor lands on the website.
The Unified Medical Language System and MetaMap
The Unified Medical Language System (UMLS) is a knowledge resource developed by the National Library of Medicine (NLM) to enable computers to understand the meaning of biomedical information14. In this study we have used the Semantic Network component in the UMLS that provides a top-level hierarchy of biomedical types such as ‘Disease or Syndrome’, ‘Anatomical Structures’ and so on called Semantic Types. The Semantic Network contains 135 Semantic Types and 54 relationships between these types. The MetaMap is a natural language processing tool developed by the NLM that maps biomedical terms/phrases to the concepts and Semantic Types in the UMLS15. The tool uses symbolic and computational linguistic techniques to perform the extraction of the relevant meaning for the term. For example, consider the term, multiple myeloma which is mapped to the Semantic Type Neoplastic Process. We use the MetaMap to process the user queries and extract the top level Semantic Types associated with the query.
Methods
Log file Analysis
We used server log files obtained from TrialX.com to understand the clinical trial information needs of online consumers. The log data were pre-processed as described below to extract the relevant query keywords that were subsequently analyzed to find semantic patterns.
1. Web Server Log Data Pre-Processing
The web server logs from TrialX.com were extracted for the period of six months.
The log entries caused by search engine crawlers and requests for static item (such as images files) were removed using an Awk (a Unix text processing utility) script.
- We extracted the following data elements from an individual log entry :
- User IP: The Internet Protocol (IP) address of the user client.
- Timestamp: The time when the web page was requested.
- Requested Webpage URL: The Uniform Resource Locator (URL) for the web page requested by the visitor.
- Referrer URL: The URL of the referring website or the search engine with request parameters.
The requested webpage URL and the referrer URL were processed to extract the user query using the appropriate GET request parameter. To illustrate, consider a requested web page URL match2trials/?keyword=glaucoma. In this request we would identify glaucoma as a user query by extracting the value for the request parameter keyword. Similarly, for a search engine, say Google, the search referrer URL would be of the form www.google.com/search?q=gastroparesis+research+texas&hl=en. We would extract the parameter ‘’ to obtain the user query ‘gastroparesis research texas’. We extract user queries using similar parameters for a Yahoo Search (p) or AOL search (query) in the referrer URL.
A unique id was assigned to unique keyword/User IP pairs and the IP addresses were purged from subsequent analysis.
2. Log Data Analysis
Keyword Frequency: The frequency of top keywords was calculated across all the queries. The frequencies were grouped based on one-word queries, two-word queries and so on. A frequency distribution of the n-word queries was plotted for n=1–10.
Semantic Patterns: The keywords were tokenized into n-gram sub-tokens, for example, gastroparesis research texas was broken down into 1-gram tokens (gastroparesis, research, texas), 2-gram (gastroparesis research, research texas and 3-gram (gastroparesis research texas). The sub-tokens were processed using the MetaMap parser (with a threshold score of 950 and above) to identify the concepts and corresponding Semantic Types. For example, using the MetaMap, gastroparesis was mapped to Disease or Syndrome, research was mapped to Research Activity and texas was mapped to Geographical Location. For each keyword, a Semantic Type vector was constructed consisting of the corresponding extracted types. The frequency of a given Semantic Type vector was calculated across all the queries.
3. User Path Evaluation
As additional analysis, we manually evaluated individual user paths through the website after the visitor landed on the website. We traced the query logs of ten randomly selected users that signed up on the website and sent a message to the trial investigator (indicating interest to enroll in the clinical trial).
Results
We found 18,429 user queries from the log data after the pre-processing step. In terms of referrers, 12,642 queries originated from google.com and tapered off subsequently with 1,091 queries from google.ca and 272 queries from search.yahoo.com. In terms of web pages requested, 13,836 clinical trial pagesφ were requested and rest were trial search result pagesη, indicating that most of the user queries are related to the text present on clinical trial pages and the user was looking for information related to clinical trials.
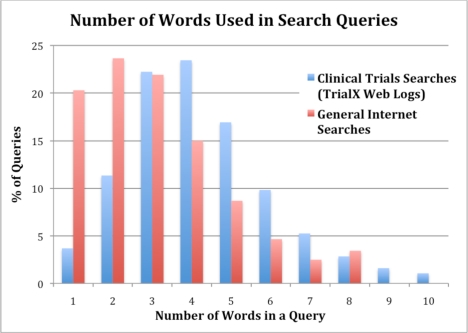
The frequency distribution based on number of words in the query is plotted in Figure 1 and compared against the queries on general Internet searches (based on data from Hitwise16). The result indicates that users looking for clinical trial information type longer queries with average of more than 3 words per query as compared to the general Internet searches. Table 1 shows the five most frequent queries for n-word long queries (for n=1 to 5). We observed that the top 1-word queries contained specific treatment or coded drug names undergoing clinical trials and the 5-word queries were more verbose where consumers were looking for trials related to a condition in a given city.
Figure 1.
The frequency distribution of the number of words per query on TrialX web logs and the general Internet searches. Clinical trials related searches on TrialX have higher average words per query (4–5).
Table 1.
Most common queries for n-words per query (n=1 to 5). The longer queries (n=4, 5) tend to contain specific information needs (location, treatment) related to clinical trials. Similarly, the top one-word queries were also specific, such as looking for drug names under the trial.
| Top 1-Word Queries | Top 2-Word Queries | Top 3-Word Queries | Top 4-Word Queries | Top 5-Word Queries | |
|---|---|---|---|---|---|
| 1 | trialx | trachelectomy procedure | obesity clinical trials | retinitis pigmentosa clinical trials | clinical trials for retinitis pigmentosa |
| 2 | vedolizumab | prochymal diabetes | osteoarthritis clinical trials | wisdom teeth study utah | clinical trials oklahoma arthritis osteoarthritis |
| 3 | hypogammaglobulin | ardsnet protocol | myelofibrosis clinical trials | permanent solution for acidity | michigan state clinical trials insomnia |
| 4 | Asp2151 | temodar melanoma | retinitis-pigmentosa clinical trials | arthritis and research studies | cystoid macular edema clinical trials |
| 5 | resvida | lucanix vaccine | als clinical trials | stretch mark clinical trial | weight loss clinical trial california |
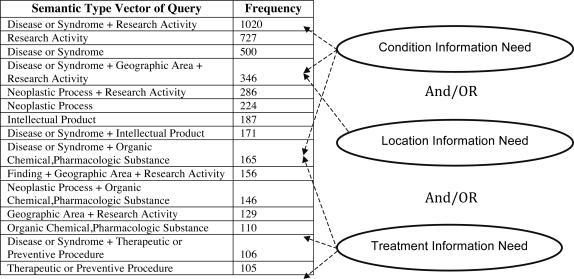
The results of Semantic Type pattern analysis are shown in Figure 2. The results indicate use of several patterns of Semantic Type combination while searching for clinical trial such as ‘Disease or Syndrome + Geographic Area’ or ‘Disease or Syndrome + Organic Chemical, Pharmacologic Substance’. We found that these patterns can be further grouped into a combination of three broad classes of information needs: condition, location, and treatment. A model of clinical trial information needs based on these classes of queries provides a substantially inclusive descriptive explanation of an online user seeking information in clinical trials, based on the data used in this study.
Figure 2.
The results of frequent combination of UMLS Semantic Types in the query keywords obtained after using MetaMap. These patterns can be grouped broadly into combination of condition, location and treatment information need. Note, the commonly used phrase ‘clinical trials’ is classified as Semantic Type Research Activity
In the manual analysis of user paths, (for users who had actually ended up sending a message to investigator), we found that 9 out of 10 search queries contained a location information need or treatment information need in addition to the condition name. All of the 10 search queries had at-least 4 or more words. One sample query had specific title of the clinical study (pilot study of mri-guided high intensity focused ultrasound ablation of uterine fibroids), while other examples contained all types of information needs in a single query (mri guided ultrasound fibroids georgia).
Discussion
The analysis presented in this study provides new and interesting insights into online information seeking in the clinical trials sub-domain. We identified a model of patient information needs in clinical trials using a semantic pattern analysis method over query logs. The results of this study provide a foundation for tailoring the online clinical trial information presented to the user.
The classes of information needs identified in the analysis are possibly unique to the clinical trials subdomain as compared to general health-related searches. An example of this is the common occurrence of location (identified as the Geographic Area Semantic Type) in the queries, indicating the importance of finding a trial close to a user’s location. Furthermore, the number of words per query for clinical trial information was found to be higher than the general Internet searches. This implies that online consumers looking for clinical trial information have a specific information need that is focused on a combination of condition, location and medication/treatment. Perhaps these are the information nuggets that need to be highlighted prominently, else such a user may bounce off the website immediately, perceiving it to not satisfy their original information need.
We believe that the types of clinical trial information needs identified in the study can be used to dynamically tailor the information presented to the user, thereby providing a richer, personalized experience on the website and potentially increase the enrollment in the studies. Consider for example, a user with the query mri-guided ultrasound uterine fibroids, which includes a condition (fibroids) and treatment (mri-guided ultrasound) information need. Hence such a user can be shown basic information about the ultrasound treatment for uterine fibroids on the top-fold of the webpage, followed by a listing of all uterine fibroid trials. Similarly, a query of mri-guided ultrasound uterine fibroids georgia includes an additional information need pertaining to a specific location. In such a case the user could be presented with a map of research site(s) conducting the relevant trials.
In this study, we looked at the query logs of a single clinical trial listing website (TrialX.com) and this represents an important limitation of this study since the results may be confounded by the ranking of TrialX web pages on search engines (this may explain the higher frequency of longer queries in our logs). Nevertheless, all the search queries analyzed were related to clinical trials since the web pages contained information only related to clinical trial protocols. Our future research includes developing dynamically constructed web pages that tailor the information based on the original consumer query and conducting bucket tests to evaluate the subsequent user paths and user engagement.
Conclusion
The consumer information needs in clinical trials have some unique characteristics and patterns. We used semantic pattern analysis over server query logs to develop a comprehensive model of consumers’ information needs for clinical trials. Understanding the broad classes of information needs can enable optimization and tailoring of clinical trial related information presented to the online consumer.
Footnotes
Work done while working previously at Applied Informatics, Inc.
Work done as a consultant for Applied Informatics, Inc.
Clinical Trial Page: http://trialx.com/clinicaltrial/88157/obesity/
Search Results: http://trialx.com/match2trials/?keyword=obesity
References
- 1.Eysenbach G, Köhler C. How do consumers search for and appraise health information on the world wide web? Qualitative study using focus groups, usability tests, and in-depth interviews. BMJ. 2002;324:573–577. doi: 10.1136/bmj.324.7337.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fox S. Online Health Search. Pew Internet & American Life Project. http://www.pewinternet.org/Reports/2006/Online-Health-Search-2006.aspx, Accessed on March 10, 2010.
- 3.ClinicalTrial.gov. http://clinicaltrials.gov/, Accessed on March 10, 2010
- 4.Sutcliffe A, Ennis M. Towards a cognitive theory of information retrieval. Interacting with Computers. 1998;10:321–51. [Google Scholar]
- 5.Keselman A, Browne AC, Kaufman DR. Consumer Health Information Seeking as Hypothesis Testing. J Am Med Inform Assoc. 2008 Jul–Aug;15(4):484–495. doi: 10.1197/jamia.M2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zeng QT, Kogan S, Plovnick RM, Crowell J, Lacroix EM, Greenes RA. Positive attitudes and failed queries: an exploration of the conundrums of consumer health information retrieval. Int J Med Inform. 2004;73:45–55. doi: 10.1016/j.ijmedinf.2003.12.015. [DOI] [PubMed] [Google Scholar]
- 7.Graham L, Tse T, Keselman A. Exploring user navigation during online health information seeking. AMIA Annu Symp Proc. 2006:299–303. [PMC free article] [PubMed] [Google Scholar]
- 8.McCray AT, Tse T. Understanding search failures in consumer health information systems. AMIA Annu Symp Proc. 2003:430–4. [PMC free article] [PubMed] [Google Scholar]
- 9.Atkinson NL, Saperstein SL, et al. Using the Internet to search for cancer clinical trials: a comparative audit of clinical trial search tools. Contemp Clin Trials. 2008 Jul;29(4):555–64. doi: 10.1016/j.cct.2008.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sillence E, Briggs P, Harris PR, Fishwick L. How do patients evaluate and make use of online health information? Soc Sci Med. 2007 May;64(9):1853–62. doi: 10.1016/j.socscimed.2007.01.012. [DOI] [PubMed] [Google Scholar]
- 11.Kreuter MW, Ricardo WJ. Tailored and Targeted Health Communication: Strategies for Enhancing Information Relevance. Am J Health Behav. 2003;27(Suppl 3):S227–S232. doi: 10.5993/ajhb.27.1.s3.6. [DOI] [PubMed] [Google Scholar]
- 12.Drennan KB. Patient recruitment: the costly and growing bottleneck in drug development. DDT. 2002 Feb;7(3) doi: 10.1016/s1359-6446(01)02137-7. [DOI] [PubMed] [Google Scholar]
- 13.Harris Interactive. 2000. www.harrisinteractive.com/news/newsletters/healthnews/HI_HealthCareNews2001Vol1_iss3.pdf, accessed Mar 10 2010.
- 14.Lindberg DA, Humphreys BL, McCray AT. The Unified Medical Language System. Methods Inf Med. 1993 Aug;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aronson AR. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Symp. 2001:17–21. [PMC free article] [PubMed] [Google Scholar]
- 16.Hitwise Data, http://image.exct.net/lib/fefc1774726706/d/1/SearchEngines_Jan09.pdf, Accessed March 14, 2010