Abstract
Many adverse drug effects (ADEs) can be attributed to drug interactions. Spontaneous reporting systems (SRS) provide a rich opportunity to detect novel post-marketed drug interaction adverse effects (DIAEs), as they include populations not well represented in clinical trials. However, their identification in SRS is nontrivial. Most existing research have addressed the statistical issues used to test or verify DIAEs, but not their identification as part of a systematic large scale database-wide mining process as discussed in this work. This paper examines the application of a highly optimized and tailored implementation of the Apriori algorithm, as well as methods addressing data quality issues, to the identification of DIAEs in FDAs SRS.
Introduction
Drug interaction adverse effects (DIAEs) are ADEs caused by special combinations of drugs, when the pharmacokinetic or pharmacodynamic properties of one drug in the combination are altered by another.
SRS are database resources encompassing reports of suspected ADEs. The availability of real-world data from SRS provides a rich opportunity to detect novel post-marketed ADEs, especially DIAEs, since in clinical trials patients on multiple drugs are usually excluded. Among the major SRSs are: FDA’s Adverse Event Reporting System (AERS), and the World Health Organization (WHO) Programme for International Drug Monitoring, which are currently the main resources in post-marketed ADE detection.
Present day SRS typically receive tens of thousands of reports each year, accumulating to this day millions of reports. Challenged by the vast size and complexity, the traditional manual case-by-case review by clinical experts, has been complemented by more efficient methods consisting of automated and quantitative approaches that are commonly referred to as data mining algorithms (DMAs). DMAs are generally designed to identify statistically strong associations between drugs and adverse effects (AEs). These associations, also referred to as signals, are not necessarily true ADEs but rather hypotheses that warrant further investigation to qualify them as credible ADEs. They allow evaluators to peruse the large volume of reports and focus their attention on potentially important safety issues.
In recent years a wide range of DMAs have been developed to screen potential ADEs1–3. The majority of DMAs rely on the use of disproportionality measures, such as the relative reporting ratio (RR)1, which attempt to quantify the degree of “unexpectedness” of a drug-AE association4. Typically, a pre-defined disproportionality threshold will be used to screen potential ADEs for further review. Both the FDA and WHO use an adjusted version of RR that accounts for the uncertainty associated with small samples, as a basis for monitoring safety signals in their SRS5, 6.
Due mostly to the voluntary nature of reporting, several issues related to data quality in SRS render the successful application of DMAs1, 7. Among the major issues identified are3, 4, 7, 8: granularity and variation in the terminology used to describe AEs/drugs, and duplicate reporting where the same ADE for the same patient is reported via different channels, or not properly linked to an earlier report as a follow-up. These data quality issues may introduce statistical biases that severely impact the results, e.g., dilution of signals across multiple similar events or drugs depending on the terminology used, or generation of spurious associations in the case of duplicate reporting.
Although some DMAs such as MGPS9 and BPCNN6 have the ability to identify ADEs attributed to multiple drugs, most studies have been designed to identify and analyze potential ADEs attributed to one drug only, e.g.,
while fewer studies focused on potential ADEs attributed to multiple drugs, e.g.,
a pharmacodynamic drug interaction where Tramadol (pain reliever) adds to the effect of Fluoxetine (Prozac) increasing serotonin levels, which may cause seizures. The importance of and difficulty of DIAE detection was emphasized in1, 7, noting that SRS databases provide an opportunity to uncover them as they contain populations that are not well represented in clinical trials. Additionally, a recent study10 examining suspected ADEs signaled by abnormal laboratory tests in hospital patients, revealed that close to 50% of the true ADEs found were due to drug interactions. By analogy, this finding suggests that many of the ADEs reported to SRS are plausibly due to drug interactions despite them being attributed to one suspected drug.
The FDA rarely receives reports attributed to drug interactions and thus their identification is not trivial. FDA’s SRS contains reports which include tens of thousands of drugs and AEs. Enumerating all possible combinations of drugs and AEs for statistical analysis is not feasible using standard approaches. This is because the search space grows exponentially with the size of the combinations considered. For example, assuming 10,000 unique drugs and AEs are under consideration, then the number of possible DIAEs due to a combination of two drugs that need to be examined is approximately 10,0003 =1012, and for each, incidence rates and other association statistics need to be computed.
Several publications in pharmacovigilance proposed methods designed to study potential DIAEs11–13. However, unlike this work, these studies all selected a small set of drugs and adverse effects prior to the mining, and addressed the statistical issues used to test or verify DIAEs. They did not address the identification of DIAEs as part of a systematic large scale mining process that is applied to the database as a whole. There are no reports of studies that mined the whole or a large subset of AERS in an attempt to identify all potential DIAEs, which we suspect is mainly due to the algorithmic complexity involved. In contrast, the work presented in this paper is on a larger scale and generality, is not confined to a specific set of drugs/AEs selected prior to mining, and focuses on the issues of mining AERS to identify all statistical associations that may correspond to potential DIAEs.
Our approach was made possible partly due to a highly optimized, parallelized, and tailored implementation of a well established data mining technique referred to as the Apriori algorithm14, that prunes the search space of possible associations.
This work builds upon a previous study we conducted15 investigating the potential use of association rules mining in SRS, but this work specifically targets DIAEs. Other major challenges addressed in this work, include: drug name mapping to generics, which is used to eliminate some of the redundancy in drug naming, thereby reducing the association space and increasing signal strength, and duplicate reporting identification and removal, which eliminates spurious associations.
Data Sources and Methods
Data Sources:
The FDA receives voluntary reports of suspected adverse drug events directly from health care professionals and consumers, as well as mandatory reports from manufacturers. Each report contains patient demographic information, drug information for as many medications as were reported for the event, including suspected drugs and concomitant drugs, coded adverse events using the MedDRA terminology (a terminology developed for ADE applications), patient outcomes, drug therapy dates, MedDRA coded indications for the reported drugs, and report sources. The data used in this work includes a large sample of individual safety reports published in 2008. This sample contained 169,040 individual reports, 24,641 unique drug names, and 8,025 unique AEs. When searching for drug interactions, we made no distinction between suspected and non-suspected medications, thus using all drugs available in each report.
Methods:
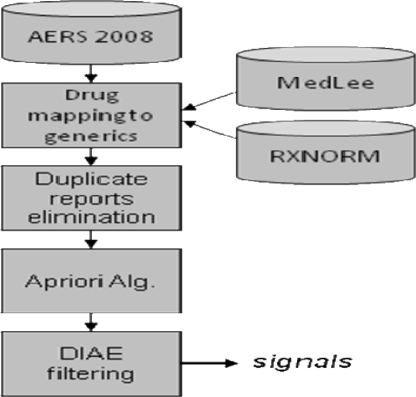
The overall mining process consisted of 4 steps depicted in Figure 1, and described in the following.
Figure 1.
Overall DIAE mining process
Drug mapping
Unlike the suspected AEs and the indications coded using MedDRA, the drugs are entered as textual terms. Each drug obtained from a report was assigned a UMLS drug code using MedLEE16, an existing NLP system. If the drug name included a dose or route, e.g. “Avandia (2 milligram) tablets”, the more general UMLS code consisting of the drug name only was chosen over the more specific code, e.g., C0875967 corresponding to “Avandia”. If the drug name could not be mapped, it was left as is. Finally, UMLS codes were mapped to generics using RXNORM. Hence, C0875967 corresponding to the brand name “Avandia”, would be mapped to C0289313 corresponding to the generic name “Rosiglitazone”.
Duplicate reporting
If not addressed properly, duplicity in reporting will produce spurious associations. E.g., for a report that contains 10 drugs and 5 AEs (a common scenario) the number of possible two drug DIAE associations will be C(10,2)×5=225, and for three drugs is 600. If this report is duplicated several times then most statistical measures of association, such as the RR measure used in this paper, will likely report these 225 (600) associations as significant and strong associations, when in fact they are spurious and only an artifact of the reporting process. Thus, if hundreds of duplicates exist, tens of thousands of spurious associations will be generated. Our method for removing duplicate reports is based on searching through reports that contain at least 8 drugs/AEs (minimizing likelihood of duplication by chance), for pairs of reports that have an exact match of the reported drugs/AEs, and demographic data (age, gender, dates). We also found that many duplicate reports did not demonstrate an exact match. This was due to missing data, e.g., gender might be included in one report but not in its duplicate, inconsistency in age, weight, and dates reported, or follow-ups which add or remove a small number of drugs/AEs. These types of duplicates were identified and removed semi-manually by repeated runs of our method. They were easy to identify as they produced highly suspicious associations with extremely large RR values.
Apriori algorithm
The Apriori algorithm is a method designed to efficiently identify association rules in large databases, and thus provides a natural setting for DIAE detection in SRS. An association rule is an implication expression of the form A→B, where A and B are disjoint itemsets. In our case, A denotes a set of drugs and B an AE, e.g., A= Fluoxetine, Tramadol, B= seizures. The strength of an association rule in the Apriori context is determined by its support and confidence, but in our case confidence is substituted by RR. The support of an itemset S(A) is the number of records containing A. The support of an association rule S(A→B) is equal to S(A ∪ B), and determines how often a rule, the combination of drugs and an AE, is observed in the data. Low support may indicate that a rule has simply occurred by chance, and thus support is one of the parameters used to eliminate uninteresting rules. The confidence of a rule which is calculated as C(A→B)=S(A ∪ B)/S(A) determines how often items in B appear in records that contain A, and provides an estimate of Pr(B|A) the conditional probability of B given A. The inappropriateness of confidence to this case stems from the fact that frequent AEs such as nausea are likely to generate large confidence values regardless of the drugs associated with it, and infrequent AEs are likely to produce small confidence despite being strongly associated with certain drugs.
In this study RR was used instead of confidence as a second parameter to qualify the worthiness or strength of an association rule. RR is defined as the ratio between a rule’s observed frequency to a baseline expected frequency, the later servings as a control. Formally, RR=N×S(A ∪ B)/S(A)S(B), where N is the total number of records in the data. RR provides an estimate of Pr(AE,Drugs)/Pr(AE)Pr(Drugs), and can therefore be viewed as the amount of deviation of the joint probability of the drug/s and AE from statistical independence. Large values indicate that the occurrence of a drug/s-AE combination has unlikely occurred by chance and that a plausible reason is behind the association. It is also easy to see that RR can be viewed as confidence normalized by the probability of B, correcting the bias discussed earlier.
The Apriori algorithm prunes the search space of associations based on the basic downward closure property of frequency. I.e, if a certain combination of drugs and AEs is infrequent, then any larger combination that builds upon the smaller infrequent one, will also be infrequent, and thus need not be considered. Despite this useful property, further optimizations were necessary in order to explore the reduced but still very large space of possible associations. These optimizations included:
Parallelization (four processors) of the major parts of the algorithm, i.e., candidate association generation, and support determination.
Hashing of reports based on drugs/AEs for more efficient support determination. Otherwise, a full scan of the database would be required to compute support for each candidate association.
Imposing the constraint that each association must include one or more drugs and one AE, thereby further reducing the space of possible associations.
The scale of computational gain achieved by these optimizations was several thousand folds, and without them the method was intractable.
In summary, our modified Apriori algorithm is used to mine the sample of AERS reports and efficiently identify associations including at least two drugs and one AE, which are later filtered by statistical measures of association to produce the final set of potential DIAEs.
Filtering
The strongest associations were filtered based on the following criteria: each association must have a support of at least 20, each association must have an RR of at least 2, a value suggested in a similar study5, and finally each association must have an RR larger than any of its subsets. The later rules out a potential DIAE that could better be explained by any of the drugs separately, and is similar to the approach proposed by Almenoff et al.13 We note that for large enough support, as in this case, an adjustment for RR that accounts for low variance as used by the FDA, would not be necessary17, 18 (adjusted and unadjusted RR are almost equal).
Evaluation
Due to lack of a gold standard (the set of all true DIAEs is unknown), evaluation in terms of sensitivity and specificity is not possible. Following common practice3, the full set of potential DIAEs identified by the method were ranked by their RR values, and a sample of 100 was given to two independent clinical experts for qualitative evaluation. Known drug interactions were validated by the experts using Micromedex and Epocrates. In addition, a hypothesis test was conducted to demonstrate that the method was not identifying drug-interactions just by chance. This was done by sampling 100 random pairs of drugs from the AERS drug distribution, and comparing the number of known drug interactions in the random sample with the number of known interactions identified by our method, using the binomial distribution. The p-value in this test is the probability of observing at least as many interactions as identified by our method, given the number expected (identified) in a random sample of 100 drug pairs.
Results
Data and association statistics: the drug mapping to generics reduced the set of drug names from 24,641 to 6,725. The number of duplicate reports found was 4094, reducing the overall set of reports from 169,040 to 163,944. Some of the reports were duplicated more than 8 times. The method produced 3402 drugs-AE associations with minimum support of 20. After filtering by RR 1868 potential DIAE remained, 1704 containing 2 drugs and 164 containing 3 drugs. We note that without duplicate report removal the method produced approximately 30,000 potential DIAEs, i.e., more than 90% spurious DIAEs! The p-value obtained for the hypothesis that our method identified drug interactions by chance was 2.8e-06.
Taxonomy of DIAEs:
Extrapolating from our evaluation sample, the full set of potential DIAEs identified by the method can be described by the taxonomy and proportions shown in Table 1. Table 2 provides representative examples of potential DIAEs classified according to the taxonomy, along with the support and RR value for each.
Table 1.
Taxonomy of potential DIAEs
| Drugs | ||
| A1 | Drugs known to be given together/ treat same indication | 57% |
| A2 | Drugs with same active ingredient | 2% |
| A3 | Supposedly unrelated drugs | 41% |
| Adverse effects | ||
| B1 | One of the drugs known to cause effect | 22% |
| B2 | All drugs known to cause effect | 21% |
| B3 | None of the drugs known to cause effect | 27% |
| B4 | Confounded association, drugs given to treat the AE | 30% |
| Interactions | ||
| C1 | Known drug interaction | 35% |
| C2 | Unknown drug interaction | 65% |
Table 2.
Classified Sample of potential DIAEs in AERS
| Taxonomy | DIAE | S | RR | |
|---|---|---|---|---|
| 1 | A1-B1-C2 | glimepiride, pioglitazone -> nausea | 20 | 2.6 |
| 2 | A1-B3-C2 | amoxicillin, clavulanate -> anaemia | 30 | 2.7 |
| 3 | A1-B4-C2 | acetaminophen, oxycodone -> back pain | 35 | 2.5 |
| 4 | A2-B2-C2 | Insulin glargine, insulin lispro -> blood glucose decreased | 40 | 26 |
| 5 | A3-B3-C1 | metformin, thyroxine -> headache | 21 | 2.8 |
| 6 | A3-B4-C1 | aspirin, furosemide -> cardiac disorder | 20 | 3.7 |
| 7 | A1-B3-C1 | digoxin, spironolactone -> drug interaction | 23 | 11 |
Discussion
The examples above illustrate various combinations of drug-drug and drug-adverse events relationships. A substantial proportion (57%) of the drugs were paired either because they were given together, e.g., amoxicillin, clavulanate (antibiotics), or because they are used to treat the same disease, e.g., glimepiride, pioglitazone treating diabetes. A small number of cases (2%), e.g., insulin glargine, insulin lispro, included different preparations containing the same active ingredient. In some cases (21%) both of the paired drugs were associated with a particular known adverse event. For example, the pair insulin glargine, insulin lispro was associated with hypoglycemia (low glucose). In the majority of drug pairs, one or none of the two drugs was known to cause the associated AE. Confounding was identified in a large number of associations (30%) where the reported AE was actually the indication for the drugs rather than a true AE. For example, the anti-diabetic agents glimepiride and pioglitazone were associated in the AERS reports with hyperglycemia, the indication for treatment, not an AE. Last, a significant proportion (35%) of drugs in a pair were known to interact with one another. For example, aspirin and furosemide were associated and known to interact since aspirin interferes with the diuretic action of furosemide. Another example is metformin which treats diabetes, and thyroxine which treats hypothyroidism; these two diseases are “autoimmune” and therefore occur together, but thyroxine decrease the effectiveness of the antidiabetic agent metformin. We also observed that in the majority of drug interactions cases where one of the drugs in the association was a known cause of the reported AE (B1–C1), the interaction was known to increase the risk of the reported AE.
As mentioned earlier, AERS rarely receives reports of drug interactions. Example 7 illustrates one of these rare cases where the corresponding reports label one of the reported AEs as “drug interaction”. In this particular case Spironolactone was labeled as the “primary suspect” drug and digoxin as the “interacting”, both known to interact. Based on the corresponding reports therapeutic agent toxicity was likely the AE attributed to the interaction, as Spironolactone may interfere with some digoxin radioimmunoassays and cause digoxin toxicity. An additional 10 DIAEs labeled as drug interactions in AERS were identified. These findings demonstrate that our method is also able to identify DIAEs that were intentionally reported as such.
Overall, the results demonstrate that a significant number of bone fide drug interactions were identified by our method. The very low p-value indicates that it is extremely unlikely that our method detected them just by chance, and thus is a valid approach. A large proportion of the associations were classified as unknown drug interactions. However, these do not necessarily indicate that the interactions do not exist, but they require further investigation. Granted, some of the unknowns can confidently be classified as spurious (drugs too long on the market for an interaction to be unknown). These typically received a very low RR value, suggesting one potential solution involving increasing the RR threshold, e.g., from 2 to 3, which could improve the filtering of spurious associations. Nonetheless, in future work we plan to investigate alternate statistical techniques to improve filtering, such as: (1) using a combination of association measures19, where the association must pass filtering by each of the measures in the combination separately (2) using logistic regression for each association to determine the statistical significance of interacting effects11.
SRS, among them AERS, may be affected by confounding1, a pathology corroborated by the findings of this study. A significant number of associations (30%) identified by our method were due to confounding, and methods to identify and remove them would be an important contribution to this field. One approach we are currently implementing is the construction of a knowledge base of known drug-disease and disease-side effect relationships, which can then be used to eliminate expected associations such as an AE which is actually a drug indication, or is closely associated with the disease treated by the drug.
We were able to remove a large proportion of duplicate reports by using an exact match approach supplemented by manual removal. However, many duplicates still remain, potentially generating a large amount of spurious associations, specifically, reports containing a small amount of drugs/AEs which we did not consider, as well as reports that would have been detected by an inexact matching approach, such as follow-ups. A possible solution based on the hit-miss model has been proposed by Noren et al.20 However, their method was fit to the WHO SRS, and it is not clear how well it will generalize to AERS. In future work we plan to explore the applicability of latter as well as new approaches to the problem.
Conclusion
A data mining technique designed to identify potential DIAEs in FDA’s AERS was presented in this paper. Our findings demonstrate its efficacy for initial identification of DIAEs. In contrast to the few existing methods, our approach is general, purely statistical, uses as much data as possible, and gives as much freedom for the data to speak for itself, without posing any restrictions or expectations on its output. Several algorithmic, as well as other challenges related to data quality were addressed in this work. Despite its relative success, several shortcomings were identified, such as confounding, and additional duplicity of reporting, which render the approach from achieving its ultimate goal, but to which solutions are currently in the works.
Acknowledgments
This research was supported in part by grants 5R01LM008635, 1R01LM010016, 3R01LM010016-01S1, and 3R01LM010016-02S1 from the National Library of Medicine.
References
- (1).Hauben M, Madigan D, Gerrits CM, Walsh L, van Puijenbroek EP. The role of data mining in pharmacovigilance. Expert Opin Drug Saf. 2005 Sep;4(5):929–48. doi: 10.1517/14740338.4.5.929. [DOI] [PubMed] [Google Scholar]
- (2).Wilson AM, Thabane L, Holbrook A. Application of data mining techniques in pharmacovigilance. Br J Clin Pharmacol. 2004 Feb;57(2):127–34. doi: 10.1046/j.1365-2125.2003.01968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (3).Almenoff J, Tonning JM, Gould AL, et al. Perspectives on the use of data mining in pharmaco-vigilance. Drug Saf. 2005;28(11):981–1007. doi: 10.2165/00002018-200528110-00002. [DOI] [PubMed] [Google Scholar]
- (4).Bate A, Evans SJ. Quantitative signal detection using spontaneous ADR reporting. Pharmacoepidemiol Drug Saf. 2009 Jun;18(6):427–36. doi: 10.1002/pds.1742. [DOI] [PubMed] [Google Scholar]
- (5).Szarfman A, Machado SG, O’Neill RT. Use of screening algorithms and computer systems to efficiently signal higher-than-expected combinations of drugs and events in the US FDA’s spontaneous reports database. Drug Saf. 2002;25(6):381–92. doi: 10.2165/00002018-200225060-00001. [DOI] [PubMed] [Google Scholar]
- (6).Bate A, Lindquist M, Edwards IR, et al. A Bayesian neural network method for adverse drug reaction signal generation. Eur J Clin Pharmacol. 1998 Jun;54(4):315–21. doi: 10.1007/s002280050466. [DOI] [PubMed] [Google Scholar]
- (7).Stephenson W, Hauben M. Data mining for signals in spontaneous reporting databases: proceed with caution. Pharmacoepidemiol Drug Saf. 2007;16(4):359–65. doi: 10.1002/pds.1323. [DOI] [PubMed] [Google Scholar]
- (8).Almenoff JS, Pattishall EN, Gibbs TG, DuMouchel W, Evans SJ, Yuen N. Novel statistical tools for monitoring the safety of marketed drugs. Clin Pharmacol Ther. 2007 Aug;82(2):157–66. doi: 10.1038/sj.clpt.6100258. [DOI] [PubMed] [Google Scholar]
- (9).DuMouchel W, Pregibon D. Empirical bayes screening for multi-item associations. Proceedings of the seventh ACM SIGKDD international conference on Knowledge discovery and data mining. 2001:67–76. [Google Scholar]
- (10).Ramirez E, Carcas AJ, Borobia AM, et al. A pharmacovigilance program from laboratory signals for the detection and reporting of serious adverse drug reactions in hospitalized patients. Clin Pharmacol Ther. 2010 Jan;87(1):74–86. doi: 10.1038/clpt.2009.185. [DOI] [PubMed] [Google Scholar]
- (11).van Puijenbroek EP, Egberts AC, Meyboom RH, Leufkens HG. Signalling possible drug-drug interactions in a spontaneous reporting system: delay of withdrawal bleeding during concomitant use of oral contraceptives and itraconazole. Br J Clin Pharmacol. 1999 Jun;47(6):689–93. doi: 10.1046/j.1365-2125.1999.00957.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (12).Bharat T, Grundschober S, Doessegger L. Detecting signals of drug–drug interactions in a spontaneous reports database. Br J Clin Pharmacol. 2007;64(4):489–95. doi: 10.1111/j.1365-2125.2007.02900.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Almenoff JS, DuMouchel W, Kindman LA, Yang X. Disproportionality analysis using empirical Bayes data mining: a tool for the evaluation of drug interactions in the post-marketing setting. Pharmacoepidemiol Drug Saf. 2003;12(6):517–21. doi: 10.1002/pds.885. [DOI] [PubMed] [Google Scholar]
- (14).Agrawal R, Imielinski T, Swami A. Mining association rules between sets of items in large databases. 1993. pp. 207–216. SIGMOD. Ref Type: Serial (Book,Monograph)
- (15).Harpaz R, Chase HS, Friedman C. Mining Multi-Item Drug Adverse Effect Associations in Spontaneous Reporting Systems. Proceedings of the AMIA Summit on Translational Bioinformatics. 2010:17. doi: 10.1186/1471-2105-11-S9-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Friedman C, Shagina L, Lussier Y, Hripcsak G. Automated encoding of clinical documents based on natural language processing. J Am Med Inform Assoc. 2004 Sep;11(5):392–402. doi: 10.1197/jamia.M1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).DuMouchel W. Bayesian data mining in large frequency tables, with an application to the FDA Spontaneous Reporting System. Am Stat. 1999;53(3):177–90. [Google Scholar]
- (18).Madigan D. Data Mining in Large Frequency Tables, with an Application to the FDA Spontaneous Reporting System: Discussion. Am Stat. 1999;53(3):198–200. [Google Scholar]
- (19).Moreno AE, Toussaint Y, Bousquet C. Mining for Adverse Drug Events with Formal Concept Analysis. Studies in Health Technology and Informatics. 2008;136:803–8. [PubMed] [Google Scholar]
- (20).Noren GN, Orre R, Bate A, Edwards IR. Duplicate detection in adverse drug reaction surveillance. Data Mining and Knowledge Discovery. 2007 Jun;14(3):305–28. [Google Scholar]