Abstract
The Unified Medical Language System (UMLS) Metathesaurus is widely used for biomedical natural language processing (NLP) tasks. In this study, we systematically analyzed UMLS Metathesaurus terms by analyzing their occurrences in over 18 million MEDLINE abstracts. Our goals were: 1. analyze the frequency and syntactic distribution of Metathesaurus terms in MEDLINE; 2. create a filtered UMLS Metathesaurus based on the MEDLINE analysis; 3. augment the UMLS Metathesaurus where each term is associated with metadata on its MEDLINE frequency and syntactic distribution statistics. After MEDLINE frequency-based filtering, the augmented UMLS Metathesaurus contains 518,835 terms and is roughly 13% of its original size. We have shown that the syntactic and frequency information is useful to identify errors in the Metathesaurus. This filtered and augmented UMLS Metathesaurus can potentially be used to improve efficiency and precision of UMLS-based information retrieval and NLP tasks.
Introduction and Background
The Unified Medical Language System (UMLS) is a project to aid the development of systems that help researchers retrieve and integrate electronic biomedical information from a variety of sources [1]. The UMLS consists of 1) a Metathesaurus which inter-connects over 100 biomedical vocabularies, 2) the Semantic Network and 3) the SPECIALIST lexicon. Of these three resources, the Metathesaurus is the most widely used resource. The 2009AB version of the UMLS Metathesaurus includes 2,120,271 biomedical concepts and 5,305,932 distinct terms from more than 100 controlled vocabularies. The UMLS Metathesaurus maps concepts among these source vocabularies. The UMLS Metathesaurus is often used as a terminology, even though it was not designed as a terminology [2].
MEDLINE is the authoritative repository of biomedical abstracts maintained by the National Library of Medicine. As of 2009, there are 19 million citations available on MEDLINE. Several information systems process the text of these abstracts with natural language processing (NLP) tools to identify concepts within the text [3].
The Metathesaurus is widely used as the underlying source for dictionary-based natural language processing (NLP) systems, such as MetaMap [4] for biomedical concept recognition and SemRep [5] for relationship extraction from the biomedical literature and from clinical documents. MetaMap—a widely used program to map concepts from the UMLS Metathesaurus to biomedical text—identifies various forms of UMLS concepts in text and returns them as a ranked list in a five-step process, which involves identifying simple NPs, generating variants of each phrase, finding matched phrases, and assigning scores to matched phrases. MetaMap’s precision estimates vary widely; Pratt et al reported a precision of 27% for MetaMap in identifying biomedical concepts from MEDLINE abstract title [6]. Shah et al. reported a precision of 9.1% when using MetaMap in identifying disease names, and a precision of 76% in identifying biological processes in MEDLINE abstracts [7].
At the National Center for Biomedical Ontology, we are developing methods to annotate large numbers of data resources automatically, and have developed an Annotator Web service for this purpose [8]. The terms recognized by the Annotator Web service come from UMLS Metathesaurus as well as the ontologies stored in BioPortal, an open repository of biomedical ontologies. The current Annotator Web service uses MGREP [9] as its concept recognizer and efforts are underway, in collaboration with NLM, to include MetaMap in the Web service. However, one of Annotator’s biggest limitations is the low precision of its underlying concept recognizers.
The goal of this study is to set the foundation for improving efficiency and precision of dictionary-based concept recognizers (e.g., MGREP and MetaMap) by analyzing the term frequency and syntactic information from 19 million MEDLINE citations. We implement our methods using UMLS Metathesaurus terms. By filtering UMLS Metathesaurus terms based on MEDLINE frequency, we can reduce the size of the lexicon allowing concept recognizers to perform more efficiently. By associating UMLS Metathesaurus terms with their syntactic statistics, we can improve the precision of the concept recognizers.
The efficiency (in terms of speed) and scalability of concept recognizers are largely determined by the size of the underlying lexicon. Shah et al. reported that in terms of speed of execution, MetaMap requires relatively long processing time, making it unsuitable for developing an online annotation service [7]. The problem of low efficiency is largely caused by the large size of the UMLS Metathesaurus. A significant number of the terms in the UMLS Metathesaurus are of little value for biomedical named entity recognition tasks and may never appear in regular text [10][11]. Examples include UMLS terms started with ‘[X]’, or terms ended with ‘, NOS’. These terms degrade the performance of applications such as MetaMap.
The precision of MetaMap is largely determined by the quality of the underlying UMLS Metathesaurus, the coverage of the SPCIALIST lexicon, and its accuracy in identifying noun phrases. Therefore, precision can be improved by creating an augmented lexicon out of the Metathesaurus.
A lexicon is a core component of any natural language processing system. The SPECIALIST lexicon is a large syntactic lexicon of biomedical and general English [12]. The SPECIALIST lexicon covers both commonly occurring English words and biomedical vocabulary. The lexicon entry for each lexical item records syntactic, morphological, and orthographic information. However, currently less than 1% of Metathesaurus terms are represented in the SPECIALIST lexicon. The SPECIALIST lexicon is created through use of an interactive lexicon-building tool, which requires significant manual effort.
There have been studies of filtering UMLS for NLP tasks. McCray et al. [11] evaluated the occurrence of UMLS Metathesaurus term in MEDLINE and constructed rules that could be used to filter out terms that are unlikely to occur naturally in a corpus. Aronson [13] has developed four filtering methods to filter out UMLS strings for MetaMap: 1. manual filtering, 2. lexical filtering, 3. filtering by type, 4. syntactic filtering. While these studies suggest that MEDLINE occurrence and string specific syntactic information are useful for filtering UMLS terms, they do not explore the corpus wide syntactic statistics for filtering out UMLS strings. The fact that the speed of MetaMap is still low after extensive filtering in MetaMap demonstrates the necessity of developing automatic approaches to systematically filter UMLS terms in order to create a view optimal for concept recognition tasks.
Data and Methods
Stanford parser is statistical parser trained on Wall Street journal [14]. The Stanford parser has been widely used in biomedical named entity recognition and relationship extraction [15][16]. We used the Stanford parser to generate syntactic information for all UMLS terms from MEDLINE abstracts. Since the Stanford parser is not a domain-specific parser, it sometimes makes mistakes in parsing biomedical text. However, the statistics gathered from a large corpus will probably crowd out the mistakes made in a single document.
18,413,784 million citations published in MEDLINE from 1965 to 2009 were parsed into sentences (96,374,837). Each sentence was syntactically parsed to generate a parse tree using the Stanford Parser. It took about 2000 core CPU days on Stanford Biox2 supercomputer cluster to parse the whole collection. We used the publicly available information retrieval library, Lucene, to create an index on sentences and their corresponding parse trees. The parse trees are available for download, search and query at http://ncbolabs-dev1.stanford.edu:8080/parsetrees.
We used UMLS 2009AB in our study, which includes 5,175,449 distinct English strings and 2,120,271 concepts. The term frequency (sentence level) was calculated by counting the occurrences of each UMLS term in all the MEDLINE sentences and abstracts. We have developed an efficient algorithm (case insensitive exact string match) for recognizing UMLS terms from MEDLINE sentences. It took about 15 minutes to map 5,175,449 UMLS terms to 96,374,837 sentences on 100 parallel computers. The syntactic types and frequencies for each term were collected from the parse trees where the term appears. Each term was then assigned a vector of syntactic types and corresponding probabilities. For example, for the term ‘breast cancer’, we have term frequency of 280,360 and the predominant syntactic type is ‘NP’ with a probability of 99.5%.
Results
Using the MEDLINE frequency and the vector of syntactic types for all the UMLS Metathesaurus terms, we develop rules to identify errors in the Metathesaurus as well as create a subset of the UMLS Metathesaurus that can improve efficiency and precision of information retrieval tasks.
1.1. Filtering based on term frequency
Table 1 shows the results of filtering based on term frequency. UMLS Metathesaurus 2009AB version contains 5,305,932 distinct English strings. Only 518,835 terms (9.1 megabytes (MB)) have ever appeared in MEDLINE, which is 13% of original terms. In addition, the size of filtered UMLS (9.2MB) is only 4% of original UMLS (221.4MB).
Table 1:
UMLS Metathesaurus terms before and after filtering
| Terms | MB | Length | Special Terms | |
|---|---|---|---|---|
| Before | 5,305,932 | 221.4 | 5 | 1,687,472 |
| After | 518,835 | 9.2 | 2 | 26,803 |
| Percent | 13.1 | 4.1 | 1.5 |
We found that the MEDLINE-based filtering predominantly filters out longer words. The average length of all UMLS Metathesaurus terms is 5 words. The average length of remaining terms after filtering is 2 words. In addition, this method filtered out words containing special characters. For example, more than 30% of original UMLS terms contain ‘, ‘, start with ‘[’, or end with ‘)’. After filtering, about 5% of UMLS Metathesaurus terms contain these special characters.
We randomly selected 100 terms that have been filtered out (terms that never appeared in MEDLINE abstracts) and evaluated them (by the first author RX). We found that the majority (95%) of them are unlikely to be used in scientific writing. This subjective evaluation suggests that term frequency based filtering effectively removes terms that are unlikely to appear in literature. The significant reduction in the number of strings (87% reduction) and space (96% reduction) will improve the efficiency of MetaMap and other UMLS Metathesaurus-based programs.
1.2. Term frequency distribution in MEDLINE
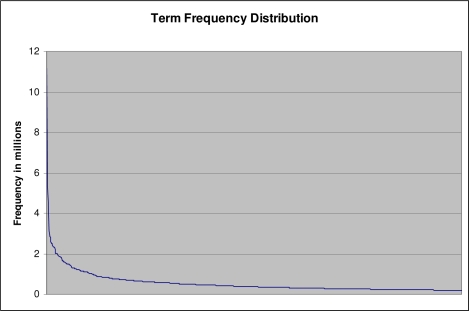
Figure 1 shows the UMLS Metathesaurus term frequency distribution in MEDLINE (data shown only for the 1000 most frequent terms). The frequency distribution of UMLS terms in MEDLINE citations generally follows the Zipf-Mandelbrot law. The most frequent term ‘patients’ appears in MEDLINE abstracts over 11 million times. 17% of UMLS Metathesaurus terms occur only once and 40% occurs fewer than five times. About 0.02% terms appear in MEDLINE abstracts more than one million times.
Figure 1:
Frequency distribution of Metathesaurus terms in MEDLINE (only top 1000 terms are shown)
We manually examined the 100 most frequent terms, and majority of them were general concepts that may not be useful for text-based concept recognition tasks. Examples of such frequent terms include ‘patients’, ‘results’, ‘disease’, and ‘drug’, which appear in MEDLINE millions of times.
Term frequency reflects the term information content. Frequent terms are often general concepts, and less frequent terms are specific concepts. The term frequency distribution provides important information for using Metathesaurus terms in text-based concept recognition. For example, the sum of MEDLINE occurrences of the top 1% terms is 40% of total occurrences of all terms. By focusing on improving quality of these highly frequent terms, the precision of UMLS-based concept recognizers can be greatly improved since more frequent terms have larger impact on the performance of concept recognizers than less frequent terms
1.3. Term distribution across SABs
Table 2 shows the top five dominant terminology sources (SABs) before and after filtering. After filtering, Medical Subject Heading (MSH) vocabulary has more terms than any other sources. This makes sense since MSH was created to index MEDLINE citations. SNOMED contains 977,316 terms and only 15% of them appear in MEDLINE. RXNORM contains 401,244 and about 2% (11,332) of them appear in MEDLINE. ICD10PCS contains 253,707 terms and only 0.008% (only two terms) ever appeared in MEDLINE abstracts.
Table 2:
Top five terminologies before and after filtering
| Order | Before Filtering | After Filtering |
|---|---|---|
| 1 | SNOMEDCT | MSH |
| 2 | MEDCIN | SNOMEDCT |
| 3 | MSH | NCBI |
| 4 | NCBI | RCD |
| 5 | RXNORM | NCI |
Our results suggest that corpus-specific filtering can be used to recommend as well as to exclude ontologies for data annotation and information retrieval tasks. For example, for MEDLINE-based information systems, MSH will be a better source ontology than either ICD10PCS or RXNORM.
1.4. Term syntactic type distribution
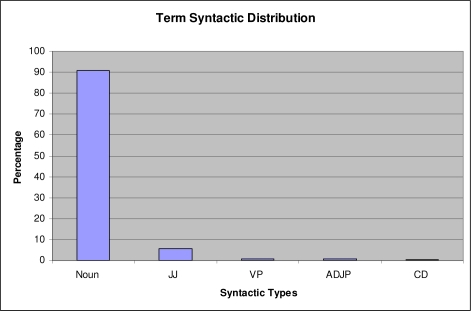
The Stanford parser was trained on a non-medical document collection and often makes mistakes in parsing biomedical documents. Therefore, instead of assigning a single syntactic type, we associate each term with a vector of syntactic types and their statistical distribution as observed over the entire corpus. For example, the Stanford parser assigned 12 syntactic types to one term ‘breast cancer’ but the dominant syntactic type is ‘NP’ (99.78%). We believe that the statistical distribution of a term’s syntactical information across a large corpus such as 19 million MEDLINE citations is more reliable than that generated from one document.
Figure 2 shows the syntactic distribution of Metathesaurus terms in MEDLINE (only the top 5 out of 36 frequent syntactic types are shown). The x-axis is the dominant type associated with a term, which is the type with highest probability. The average number of syntactic types for each term analyzed is 3.5. In the example of “breast cancer”, the dominant syntactic type is ‘NP’ with probability of 99.78%. As shown in the figure, over 90% of UMLS Metathesaurus terms have the dominant syntactic type of noun phrase (e.g., NP, NN, NNS, NNP, NNPS), and 5% have dominant syntactic type of ‘JJ’ (adjective).
Figure 2:
Syntactic distribution of terms in MEDLINE (top 5 syntactic types out of 36 are shown). The most common syntactic type is noun phrase.
We manually examined the top 100 terms with the dominant syntactic type of “NP”, and all of them are true noun phrases. We then manually checked the top 100 terms whose dominant types are not noun phrase. All of them are indeed not noun phrases. This shows that even though the Stanford parser makes mistakes in parsing individual sentences from biomedical text (as shown by the number of syntactic types associated with each term), the dominant syntactic types over the entire MEDLINE distribution statistics are likely to be correct. However, as the frequency decreases, the syntactic type statistics will be less certain.
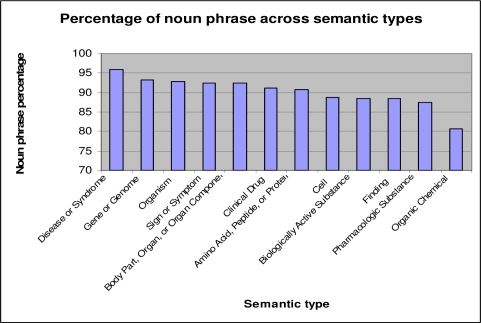
1.5. NP distribution across semantic types
Figure 2 shows the percentage of noun phrase in 12 selected semantic types from the UMLS Semantic Network. For example, over 95% of terms with semantic type ‘Disease or Syndrome’ have dominant syntactic type of noun phrase. We manually checked the top 100 frequent terms with semantic type of ‘Disease or Syndrome’ and with syntactic types other than noun phrase. We found that 95% of them are indeed not noun phrases. For example, terms such as ‘renal’, ‘little’, ‘best’ and ‘infectious’ have been assigned semantic type ‘Disease or Syndrome’, but are not noun phrases. Frequent terms, if mistyped, will hurt the precision of NLP-based concept recognizers significantly. This example shows that we can automatically flag incorrectly typed terms based on the syntactic type distribution, and, by removing these incorrect terms, we can improve the precision of NLP-based concept recognition systems.
Discussion
We have developed a method to filter the UMLS Metathesaurus by analyzing MEDLINE abstracts. The filtered UMLS Metathesaurus is 13% of original size. We have augmented the UMLS Metathesaurus by incorporating each term’s frequency and syntactic distribution statistics from MEDLINE. We have argued that by incorporating the MEDLINE syntactic distribution statistics, we can improve the quality of lexicons derived out of the Metathesaurus and improve the precision of dictionary-based concept recognizers.
The distribution of UMLS Metathesaurus terms in MEDLINE is skewed. About 15% of terms appear only once and 27% of terms at most twice. The syntactic information for terms with such low frequency may not be as reliable as that of common terms. It might be best to assume that these terms are all noun phrases and to ignore the syntactic types assigned by the parser. Since these are rare terms and account for only 0.00000071% of total UMLS Metathesaurus term occurrences, this assumption will have minimal impact on the performance of dictionary-based concept recognizers.
There are 220 ontologies in NCBO’s BioPortal, with about 8 million term names. Without filtering, it is hard to use these terms in a dictionary with concept recognizers such as MetaMap. In addition, there exists no lexicon—analogous to the SPECIALIST lexicon—which has extensive syntactic information for these terms. The methods we have developed for Metathesaurus terms are directly applicable to the terms from the ontologies stored in BioPortal for improving the precision and efficiency of ontology-based concept recognizers.
Since most biomedical concepts are noun phrases, we can improve the quality of lexicons derived from the UMLS Metathesaurus or BioPortal ontologies by removing those terms whose dominant syntactic types are not noun phrases. In addition, by focusing on removing the most frequent terms, we expect a large improvement in precision of ontology-based concept recognizers. For example, the 100 most frequent terms account for 19% of the sum of all occurrences of UMLS Metathesaurus terms in MEDLINE. Most of the common terms, such as ‘study’, ‘treatment’, ‘patients’ or ‘results’, have little value for ontology-based concept recognizers.
Currently, the term frequency and syntactic information are collected from MEDLINE abstracts. It is very likely that the term information distribution in clinical documents or descriptions of gene expression data-sets is different from that in MELDINE. Because of the noisy nature of clinical documents, it will be interesting to see whether or not these methods (especially the syntactic parsing method) can be used to generate corpus-specific statistics of term frequency and syntactic statistics in order to create custom lexicons to be used in clinical corpora.
Figure 3:
Distribution of noun phrases across semantic types from the UMLS semantic network (12 out of 135 semantic types are shown).
Acknowledgments
This work was funded by the National Center for Biomedical Ontology under NIH grant U54 HG004028.
References
- 1.Lindberg DA, Humphreys BL, McCray AT. The unified medical language system. Methods Inf Med. 1993 Aug;32(4):281–91. doi: 10.1055/s-0038-1634945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Perl Y, Geller J, Cimino J. Analysis of a study of the users, uses and future agenda of the UMLS. JAMIA. 2007;14(2) doi: 10.1197/jamia.M2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Srinivasan S, Rindflesch TC, Hole WT, Aronson AR, Mork JG. Finding UMLS Metathesaurus concepts in MEDLINE. In Proc AMIA Symp. 2002 [PMC free article] [PubMed] [Google Scholar]
- 4.Aronson AR. Effective mapping of biomedical text to the UMLS metathesaurus: The metamap program. In Proc of AMIA Symp. 2001 [PMC free article] [PubMed] [Google Scholar]
- 5.Rindflesch TC, Fiszman M. The interaction of domain knowledge and linguistic structure in natural language processing: interpreting hypernymic propositions in biomedical text. J of Biomed Inform. 2003 doi: 10.1016/j.jbi.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 6.Pratt W, Yetisgen-Yildiz M. A study of biomedical concept identification Metamap vs. people. In Proc AMIA Symp. 2003 [PMC free article] [PubMed] [Google Scholar]
- 7.Shah NH, Bhatia N, Jonquet C, Rubin D, Chiang AP, Musen MA. Comparison of concept recognizers for building the open biomedical annotator. BMC Bioinformatics. 2009;10(Suppl 9):S14. doi: 10.1186/1471-2105-10-S9-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jonquet C, Shah NH, Musen MA. AMIA Summit on Translational Bioinformatics. San Francisco: 2009. The open biomedical annotator. [PMC free article] [PubMed] [Google Scholar]
- 9.Dai M, et al. AMIA Summit on Translational Bioinformatics. San Francisco, CA: 2008. An efficient solution for mapping free text to ontology terms. [Google Scholar]
- 10.Roberts A, Gaizauskas R, Hepple M, Guo Y. Combining terminology resources and statistical methods for entity recognition: an evaluation. In proceedings of the sixth international conference on language resources and evaluation, LREC; 2008. [Google Scholar]
- 11.McCray A, Bodenreider O, Malley J, Browne A. Evaluating UMLS Strings for Natural Language. In Proc of AMIA Symp. 20012001 [PMC free article] [PubMed] [Google Scholar]
- 12.McCray AT, Srinivasan S, Browne AC. Lexical methods for managing variation in biomedical terminologies. Proc Annu Symp Comput Appl Med Care. 1994:235–9. [PMC free article] [PubMed] [Google Scholar]
- 13.Aronson A. Filtering the umls metathesaurus for metamap. 2009. Technical report, U.S. National Library of Medicine, Lister Hill National Center for Biomedical Communications.
- 14.Manning CD, Klein D. Accurate unlexicalized parsing; In Proc of the 41st Meeting of the Association for Computational Linguistics; 2003. [Google Scholar]
- 15.Xu R, Supekar K, Morgan A, Das A, Garber A. Unsupervised method for automatic construction of a disease dictionary from a large free text collection. In Proc of AMIA Symp. 2008 [PMC free article] [PubMed] [Google Scholar]
- 16.Xu R, Das A, Garber A. Unsupervised method for extracting machine understandable medical knowledge from a large free text collection. Proc of AMIA Symp. 2009 [PMC free article] [PubMed] [Google Scholar]