Abstract
Accurate assessment and evaluation of medical curricula has long been a goal of medical educators. Current methods rely on manually-entered keywords and trainee-recorded logs of case exposure. In this study, we used natural language processing to compare the clinical content coverage in a four-year medical curriculum to the electronic medical record notes written by clinical trainees. The content coverage was compared for each of 25 agreed-upon core clinical problems (CCPs) and seven categories of infectious diseases. Most CCPs were covered in both corpora. Lecture curricula more frequently represented rare curricula, and several areas of low content coverage were identified, primarily related to outpatient complaints. Such methods may prove useful for future curriculum evaluations and revisions.
Introduction
Educators have long sought to tailor curricula to the needs of their students. Governing bodies such as the American Association of Medical College’s (AAMC) Liaison Committee for Medical Education (LCME) accredit medical schools on prescribed curricula, and are increasingly mandating more detailed accounting for the content taught and clinical experiences during medical school. These efforts are based largely on expert panel recommendations and are implemented at local levels through curriculum committees and individual course directors. Yet no quantitative methods to match topics taught to topics learned yet exist. In this paper, we compare the concepts taught in the four-year curriculum to those written about in clinical notes. We apply natural language processing tools to a comprehensive curriculum management system and electronic medical record notes to quantify differences between each.
The current state-of-the-art at most institutions is categorical assessments of “keywords” covered in the curricula. To allow such cataloging of a school’s curriculum, the AAMC has created the Curriculum Management and Information Tool (CurrMIT), which allows manual entry of lectures, teachers, keywords, and topics covered.1 Such data can be compared across different institutions,2,3 and represents a data source often used for LCME visits. Other programs, such as Tapestry and the Tufts University Sciences Knowledgebase, have created systems to allow for more efficient collection of this metadata. These metadata are limited in their ability to describe the depth of concepts discussed, are not amenable to unforeseen search queries (since content areas are typically specified a priori), and require significant manual effort to document.
At Vanderbilt, we have addressed these needs through the creation of a concept-based curriculum management program called KnowledgeMap (KM).4 KM has been used for curriculum management since 2002 and supports undergraduate and graduate medical education. The heart of KM is the KM concept indexer (KMCI), a robust natural NLP tool that identifies Unified Medical Language System (UMLS) concepts from text. KMCI has been described previously;5 it uses a rigorous, score-based method to disambiguate unclear document phrases using contextual clues. Faculty upload curricular materials in a variety of formats, such as Microsoft PowerPoint®; these are converted into text documents and parsed by KMCI. Faculty can search for content through concept-based searches, find related PubMed articles, and perform broad queries of curricula on topics such as “genetics” or “geriatrics”.6
As trainees write clinical notes in the medical record, they are captured by the Learning Portfolio (LP) system.7 LP uses an NLP tool called SecTag8,9 to identify note sections (e.g., “history of present illness”, “cardiovascular exam”) followed by KMCI to allow educators to answer such questions as whether a student performed a straight leg raise on the patients he/she saw with back pain. LP was introduced in 2005 with medical students and expanded to include most housestaff physicians in 2007. Currently, LP is used to enhance feedback between students and mentors,7 keeping logs of procedures performed, and tracking their clinical exposure.10
Led by a team of associate deans and master clinical educators, the Vanderbilt School of Medicine has prioritized 25 core clinical problems (CCP) to be mastered by graduating medical students; collectively, these are called the “Core Clinical Curriculum”.11 Each CCP addresses a common patient presentation that ranges from serious illnesses to every-day complaints. For each of the 25 problems, the team developed a set of learning objectives that included 30–60 descriptive elements for each. The representative objectives include specific history items, physical exam findings, differential diagnoses and appropriate diagnostic evaluation that a finishing medical student should have learned. For example, the topic of back pain includes the representative concepts “history of cancer,” “straight leg raise exam,” and “spinal cord compression.” Students are currently tracking their clinical experiences according to these 25 topics, and meeting periodically with senior educators to review their progress toward competency.
In this paper, we provide a first step toward addressing the challenge of matching what is taught to what is experienced. As a proof-of-concept, we compared the distribution of infection and CCP-related concepts in clinical notes and curriculum documents to evaluate the ability of this method to elucidate areas of the curriculum potentially needing further development.
Methods
Use of KMCI in KM and LP produce lists of UMLS concepts that are stored in relational databases, linked to their original document. In LP, these concepts are also categorized by the section (e.g., “history of present illness”) to which they belong. Documents in KM are indexed by their curriculum year. We identified two areas to compare content coverage, infectious diseases and CCPs. These were chosen by educators as two content categorizations of local interest and important for preparation for LCME criteria. In this study, we compared the content coverage of infectious diseases and CCPs in 2008–2009 curricular documents from KM (“KMD”) to all medical trainee-authored clinical notes in LP since 2005 (“LPN”).
Creating queries for CCP content:
The project team, led by author AS with significant input with local content experts and senior medical educators, created broad sets of UMLS concepts defining search strategies for each of the 25 CCPs. CCP concept lists began with free-text “learning objective” documents describing each CCP. Each CCP document was created using a structured template consisting of goals for medical knowledge; key history, physical examination maneuvers, and tests the student should consider; initial workup for the condition; and the differential diagnosis, by system, of the presenting problem. Each CCP objective document was revised by several senior medical educators.
The final version of each CCP document was placed on KM and processed with KMCI to generate a list of UMLS concepts for each CCP documents. Authors AS and JD expanded key concepts, such as “abdominal pain”, using a tool in KM that allows one to add and refine searches with related UMLS concepts (e.g., “right upper quadrant pain”).6 Finally, concept lists were expanded by finding related curricular documents in KM (by searching for such keywords as “abdominal pain” or “dysuria”) and then adding additional concepts judged relevant by content experts or the project team. Educators were assisted in this process by a display of UMLS concepts in each document, sorted by term weight (based on a term-frequency, inverse document frequency model12) and frequency.
When finalized, each search strategy consisted of between 19 and 532 unique concepts (i.e., concept unique identifiers, or CUIs). Ten of these concept queries were formally evaluated in a previous study, which compared multiple scoring algorithms to best identify which clinical notes discussed each topic.10 The concept lists were revised iteratively via application in the curriculum, adding related concepts and removing others that were less important.
Identifying infectious diseases:
We identified seven categories of infectious diseases to catalog: bacteria, virii, fungi, protozoans, helminths, prions, and tick-borne diseases. Bacteria, Virii, and Fungi were identified by selecting all concepts with those respective UMLS semantic types. Protozoans, helminths, and tick-borne diseases were identified by finding all UMLS concepts of related to these “parent” concepts with UMLS-specified relationships of child (CHD), related-like (RL), related-narrow (RN), and synonymous (SY). Concept lists were built and edited using the KM interface6 by a clinician-author (JD). For each grouping, the concept lists contained the following counts of unique concepts (i.e., CUIs): bacteria – 88,898, virii – 7,109, fungi – 39,100, protozoans – 2387, helminths – 1451, tick-borne diseases – 247, and prions – 21.
Calculation of concept coverage:
We were primarily interested in identifying the amount of content in the curricula and clinical notes devoted to each topic. Thus, we extracted the lists of UMLS concepts matching each CCP from the KM and LP concept databases. We limited our search of KM to all documents contained within the last completed curriculum year (2008–2009), which includes all required courses through the four year curriculum (28 courses). To achieve a greater collection of rare concepts (diseases and infections), we included all clinical notes in LP. We included both medical students and housestaff physician-generated notes (primarily pediatric and internal medicine residencies at this time), since the goal of medical school is to train a student for clinical medicine, not just the third and fourth year of medical school. The study was performed using the 2009AA edition of the UMLS.
For each concept list, we calculated the “concept density” of that topic in each corpus (e.g., KMD or LPN). For each CCP, the concept density was calculated as the ratio of the number of concepts matching the CCP concept list to the number of concepts matching any CCP in that corpus. Similarly, the concept density for infections was calculated as the ratio of number of concepts matching a specific infectious disease class (e.g., virii) to the number of concepts matching any infectious disease.
Results
Table 1 presents the overview of the KMD and LPN corpora. Understandably, there were many more clinical notes than curricular documents, and they covered a broader range of unique concepts than the curriculum documents. Curricular documents tended to have more total and unique concepts than clinical notes.
Table 1.
Overview of KMD and LPN corpora.
| Clinical Notes | Curriculum Documents | |
|---|---|---|
| Count | 339,518 | 2049 |
| Unique concepts | 139,989 | 69,490 |
| No. concepts | 130,608,924 | 1,471,447 |
| No. relevant concepts | 15,586,342 | 111,042 |
| Median concepts/document (IQR) | 338 (213–526) | 558 (203–1012) |
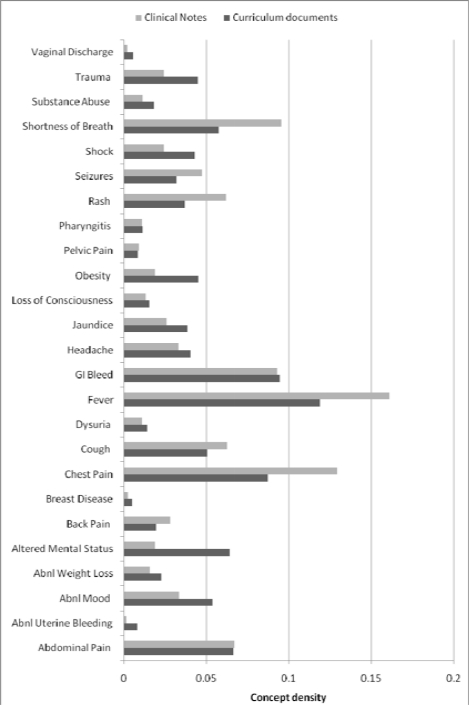
Figure 1 presents the concept density for all CCPs. The most common CCPs covered in clinical notes were fever, chest pain, shortness of breath, and GI bleeding. The most common CCPs covered in curricular documents were similar. Fever, chest pain, and altered mental status were the most discrepant CCPs between KMD and LPN. Several gynecological CCPs (breast disease, abnormal uterine bleeding) were rarely covered in either corpus.
Figure 1.
Concept density comparison for 25 core clinical problems.
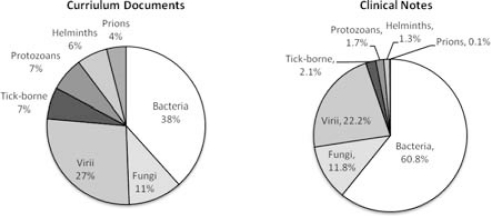
Figure 2 presents the distribution of infectious diseases between the corpora; KMD contained a much greater discussion of prions, helminths, protozoans, and tick-borne diseases, but a significantly less discussion of bacteria. Of note, 516 unique bacteria and 56 fungi were discussed in LPN not mentioned in KMD; most of these were specific species of genus otherwise mentioned in KMD.
Figure 2:
Concept density comparison for infectious disease coverage.
Discussion
The assessment and revision of the medical curriculum remains challenging. We assessed the content coverage of two components of clinical training at an academic medical school: the four-year didactic curriculum and the clinical experiences documented through notes produced by its trainees. Our analyses demonstrate that the majority of the core clinical topics represented by CCPs were covered by both corpora in similar ratios. Finally, the method allows rapid assessment of concepts covered in one source not discussed in another. To our knowledge, this study is the first to apply NLP methods to automatically categorize and evaluate curricula and clinical exposure according to an a priori established rubric. Such a method may serve to assist in future curriculum evaluation and revision.
The method highlighted several areas of low clinical exposure (e.g., LPN). Several CCPs representing common problems (vaginal discharge, breast disease, and abnormal uterine bleeding) were rare in clinical notes; this likely represents the bias of our clinical training toward inpatient experiences. Discrepancies, such as seen with the obesity, may indicate a clinical trainees’ lack of discussion of obesity-related concepts in obese patients, given the high prevalence of obesity. Such knowledge may highlight areas for focused intervention in the clinical curriculum.
Determining the right “dose” and “breadth” of medical content to teach to medical trainees is challenging. Identifying concepts taught in medical school that are rarely (if ever) seen in clinical notes does not indicate that the concept does not need to be taught. Medical education needs to expose students to broad categories of knowledge, including rare diseases (e.g., prion diseases) and diseases not common in the United States (e.g., malaria). Thus, one expects to find a broader coverage of concepts in a curriculum than in clinical notes. In comparison, clinical notes, however, contained many specific bacteria that are never discussed in curriculum documents. In the vast majority of the cases, these bacteria are discussed at the genus level. Didactic teaching of bacteria and their treatments are more important given the prevalence of bacterial infections in the hospital and clinic settings. Future work should consider the clinical importance of concept coverage differences and concepts present in one corpus not found in another, potentially using UMLS-specified relationships to automate this task.
Several limitations caution interpretation of these data. Our lists of concepts were automatically derived, using KMCI. While KMCI and SecTag have performed well in related studies5–10, in a corpus this large, even small error rates will generate a significant number of false positive individual concept identifications. Moreover, systematic errors are possible that ignore large quantities of data. However, it is infeasible to manually curate such a large set. Second, our capture of clinical experience relies on those notes written at Vanderbilt Medical Center (VMC). While the majority of training is performed at VMC and nearly all notes are electronic, some content is missed, such as training in outside hospitals and in programs for which clinical documentation may not be a major aim of the rotation (e.g., general surgery). We have attempted to correct for the latter concern by including housestaff notes as well. Finally, the list of concepts for each CCP may contain a bias toward content already present in the curriculum, since the search strategies were created in part from the concepts already present in key curricular documents. However, this bias would echo the mental “filters” employed by educators as they decide what content to teach.
Our previous experience with automatically-generated procedure counts suggests that this automated capture will far outpace that captured by manual logging. Thus, we believe methods such as presented in this study may provide a more accurate accounting of the curriculum and clinical experience than traditional, labor-intensive methods.
References
- 1.Salas AA, Anderson MB, LaCourse L, et al. CurrMIT: a tool for managing medical school curricula. Acad Med. 2003;78(3):275–279. doi: 10.1097/00001888-200303000-00009. [DOI] [PubMed] [Google Scholar]
- 2.Henrich JB, Viscoli CM. What do medical schools teach about women’s health and gender differences? Acad Med. 2006;81(5):476–482. doi: 10.1097/01.ACM.0000222268.60211.fc. [DOI] [PubMed] [Google Scholar]
- 3.Van Aalst-Cohen ES, Riggs R, Byock IR. Palliative care in medical school curricula: a survey of United States medical schools. J Palliat Med. 2008;11(9):1200–1202. doi: 10.1089/jpm.2008.0118. [DOI] [PubMed] [Google Scholar]
- 4.Denny JC, Irani PR, Wehbe FH, Smithers JD, Spickard A. The KnowledgeMap project: development of a concept-based medical school curriculum database. AMIA Annu Symp Proc. 2003:195–9. [PMC free article] [PubMed] [Google Scholar]
- 5.Denny JC, Smithers JD, Miller RA, Spickard A. “Understanding” medical school curriculum content using KnowledgeMap. J Am Med Inform Assoc. 2003;10(4):351–62. doi: 10.1197/jamia.M1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Denny JC, Smithers JD, Armstrong B, Spickard A. “Where do we teach what?” Finding broad concepts in the medical school curriculum. J Gen Intern Med. 2005;20(10):943–6. doi: 10.1111/j.1525-1497.2005.0203.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spickard A, Gigante J, Stein G, Denny JC. Automatic capture of student notes to augment mentor feedback and student performance on patient write-ups. Journal of general internal medicine. 2008;23(7):979–84. doi: 10.1007/s11606-008-0608-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Denny JC, Spickard A, Johnson KB, et al. Evaluation of a method to identify and categorize section headers in clinical documents. J Am Med Inform Assoc. 2009;16(6):806–815. doi: 10.1197/jamia.M3037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Denny J, Miller R, Johnson K, Spickard I. Development and Evaluation of a Clinical Note Section Header Terminology. Proc AMIA Symp. 2008:156–60. [PMC free article] [PubMed] [Google Scholar]
- 10.Denny JC, Bastarache L, Sastre EA, Spickard A. Tracking medical students’ clinical experiences using natural language processing. J Biomed Inform. 2009;42(5):781–789. doi: 10.1016/j.jbi.2009.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gotterer GS, Petrusa E, Gabbe SG, Miller BM. A program to enhance competence in clinical transaction skills. Acad Med. 2009;84(7):838–843. doi: 10.1097/ACM.0b013e3181a81e38. [DOI] [PubMed] [Google Scholar]
- 12.Tf-idf. Available at: http://en.wikipedia.org/wiki/Tf-idf [Accessed October 27, 2009].