Abstract
For many clinical conditions, only a small number of patients experience adverse outcomes. Developing risk stratification algorithms for these conditions typically requires collecting large volumes of data to capture enough positive and negative for training. This process is slow, expensive, and may not be appropriate for new phenomena. In this paper, we explore different anomaly detection approaches to identify high-risk patients as cases that lie in sparse regions of the feature space. We study three broad categories of anomaly detection methods: classification-based, nearest neighbor-based, and clustering-based techniques. When evaluated on data from the National Surgical Quality Improvement Program (NSQIP), these methods were able to successfully identify patients at an elevated risk of mortality and rare morbidities following inpatient surgical procedures.
Introduction
For many clinical conditions, patients experiencing adverse outcomes comprise a small minority in the population. For example, the rate of many important clinical complications, ranging from coma to bleeding, was well below 1% in the National Surgical Quality Improvement Program (NSQIP) data sampled at over 200 hospital sites1. Roughly 2% of patients undergoing surgery at these sites died in the 30 days following the procedure.
Identifying patients at risk of adverse outcomes in such a setting is challenging. Most existing algorithms for clinical risk stratification rely on the availability of positively and negatively labeled training data. For outcomes that are rare in a population, using these algorithms requires collecting data from a large number of patients to capture a sufficient number of positive examples for training. This process is slow, expensive, and often imposes burden on caregivers and patients. The costs and complexity of extensive expertly-labeled data points have impeded the spread of even well validated and effective health care quality interventions2.
Our recent research has focused on detecting high-risk patients in a population as anomalies3. Data from our earlier studies shows that this approach can identify patients at risk of both adverse cardiac and critical care outcomes. In some cases, identifying patients as anomalies can even outperform supervised methods that use additional knowledge in the form of positive and negative examples3. This is due to supervised methods being unable to generalize for complex, multi-factorial events where only a few patients in a large training set experience events.
In this paper, we improve upon our earlier work by investigating the relative merits of different anomaly detection methodologies to identify high-risk patients without the need of labeled training examples. In the absence of any expert knowledge, these methods search for cases that lie in sparse regions of the feature space. We explore three different categories of unsupervised anomaly detection methods: classification-based, nearest neighbor-based, and clustering-based techniques. When evaluated on data from over 100,000 patients undergoing inpatient surgical procedures, all three approaches were able to successfully identify patients at increased risk of mortality and rate morbidities.
Background
Most existing work on prognosticating patients relies on the availability of positively and negatively labeled examples for training. In contrast to these methods, which attempt to develop models for individual diseases, we focus on unsupervised anomaly detection to identify high-risk patients. The hypothesis underlying our work is that patients who differ the most from other patients in a population are likely to be at an increased risk of adverse outcomes.
Anomaly detection has been studied in the context of medicine in earlier work. Closely related to our research are efforts to use anomaly detection to evaluate patient data. For example, Tarassenko et al.4 applied novelty detection to masses in mammograms, Campbell and Bennett5 to blood samples, Roberts and Tarassenko6 to electroencephalographic signals, and Laurikkala et al.7 to vestibular data. Previous work has focused on clinician behavior or specific technical testing parameters, yet has avoided the realm of quality and outcomes in health care in an important and relevant way that could help us improve patient care.
We add to these efforts by developing and evaluating the use of unsupervised anomaly detection more broadly. In contrast to previous work focusing on detecting existing disease, our research uses anomaly detection to identify patients at increased risk of adverse future outcomes. We describe how different approaches can be used to achieve this goal, and compare these methods in a real-world clinical application. As part of this process, we evaluate unsupervised anomaly detection on a larger, more broadly clinically relevant dataset than any of the earlier studies (both in terms of number of patients and different clinical outcomes studied).
Methods
Classification-Based Anomaly Detection
Our first approach to identify patients who are anomalies in a population is based on one-class support vector machines (SVMs). Given the normalized feature vectors xi for patients i=1,...,n, we develop a one-class SVM using the method proposed by Scholkopf et al.8 This method first maps the data into a second feature space F using a feature map Φ. Dot products in F can be computed using a simple kernel
such as the radial basis function (RBF) kernel:
In contrast to the standard SVM algorithm9, which separates two classes in the feature space F by a hyperplane, the one-class SVM attempts to separate the entire dataset from the origin. This is done by solving the following quardratic problem (which penalizes feature vectors not separated from the origin, while simultaneously trying to maximize the distance of this hyperplane from the origin):
subject to:
where v reflects the tradeoff between incorporating outliers and minimizing the support region size.
The resulting decision function, in terms of the Lagrange multipliers, is then:
We detect anomalies in a population by first developing a one-class SVM on the data for all patients, and then using the resulting decision function to identify patients with feature vectors that are not separated from the hyperplane. These patients, who lie outside the enclosing boundary, are labeled as anomalies. The parameter v can be varied during the process of developing the one-class SVM to control the size of this group.
Nearest Neighbor-Based Anomaly Detection
Our second approach to identify patients who are anomalies in a population is based on k-nearest neighbors. We use an approach similar to that of Eskin et al.10 to find feature vectors that occur in dense neighborhoods. This is done by computing the sum of the distances from each normalized feature vector to its k-nearest neighbors (i.e., the k-NN score for each point in the feature space). Points in dense regions will have many points near them and will have a small k-NN score. Conversely, points that are anomalies will have a high k-NN score.
The computational runtime of having to find the k-nearest neighbors of each point can be decreased by partitioning the space into smaller subsets so as to remove the necessity of checking every data point11.
Cluster-Based Anomaly Detection
Our third approach to identify patients who are anomalies in a population is based on cluster-based estimation10. For each feature vector xi, we define N(xi) to be the number of points within W of xi:
These values can be used to estimate how many points are “near” each point in the feature space. Patients with low values can be identified as anomalies.
We use a fixed width clustering algorithm to approximate N(xi) in a computationally efficient manner for all i=1,…,n. This algorithm sets the first point as the center of the first cluster. Every subsequent point that is within W of this point is added to the cluster, or is otherwise set to be a new cluster. Some points may be added to multiple clusters using this approach. At the end of this process, N(xi) is approximated by the number of points in the cluster to which xi belongs.
Evaluation
We evaluated our work on data from 178,491 patients undergoing inpatient surgical procedures in the 2008 NSQIP Participant Use File (PUF). These data were sampled at 203 hospital sites and consisted of patients undergoing both general and vascular surgery. Patients were followed for 30 days post-surgery for mortality and various morbidities.
The features for each patient consisted of 45 preoperative demographic and comorbidity variables. We did not make use of laboratory results as these values were missing in a large fraction of the patients. A total of 137,774 patients did not have any missing data for the demographic and comorbidity variables used.
We studied the ability of the different anomaly detection methods to risk stratify these patients for 10 rare morbidity outcomes: peripheral nerve injury (124 events), coma >24 hours (176), myocardial infarction (312), failure of extracardiac graft or prosthesis (483), stroke or cerebrovascular accident (508), pulmonary embolism (634), renal insufficiency (693), cardiac arrest (783), bleeding requiring transfusion (847), and renal failure (881). We also studied the endpoint of mortality (2,997).
We developed the one-class SVM using LIBSVM (National Taiwan University). Given the focus of our work on unsupervised anomaly detection we used default parameter choices in LIBSVM (i.e., RBF kernel with γ set to 1/number of features) with v set to obtain an anomaly group comprising patients lying in the most dissimilar quintile relative to the rest of the population. For the k-nearest neighbor and cluster-based estimation approaches, we used MATLAB (Mathworks Inc.) implementations. The value of k was set to 3 for the k-nearest neighbor approach, with patients having k-NN scores in the highest quintile being classified as being anomalies. For cluster-based estimation, we set W=4 and identified patients in the lowest quintile of N(x) values as anomalies. For each anomaly detection method, we used a single model developed as described above to predict all outcomes.
We assessed the predictive ability of unsupervised anomaly detection in two ways. First, we studied the categorization of patients by all three methods using Kaplan-Meier survival analysis to compare event rates between anomaly and non-anomaly groups. Hazard ratios (HR) and 95% confidence intervals (CI) were estimated using a Cox proportional hazards regression model. Consistent with the medical literature, findings were considered to be significant for a p-value less than 0.05. We also assessed the predictive ability of unsupervised anomaly detection by calculating the area under the receiver operating characteristic curve (AUROC) for each anomaly detection approach. All statistical analyses were conducted in MATLAB. This work was done with the approval of the Henry Ford Health System Institutional Review Board and under the Data Use Agreement of the American College of Surgeons.
Results
Table 1 presents the hazard ratios of patients categorized as being anomalies by each of the three unsupervised anomaly detection approaches. Patients identified as anomalies by each method showed an elevated risk for all but one morbidity endpoint (peripheral nerve injury) and for the outcome of death within 30 days of inpatient surgery. Most morbidities showed a 2- to 3-fold increased risk of adverse outcome. This increase in risk was even higher for death.
Table 1:
Hazard ratios (HR), 95% confidence intervals (CI) and p-values (P) of all three unsupervised anomaly detection approaches for different morbidities and the endpoint of mortality within 30 days of inpatient surgery. There were a total of 137,774 patients with each anomaly detection approach identifying a quintile as anomalies.
| One-Class Support Vector Machine | k-Nearest Neighbor | Cluster-Based Estimation | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Event % | HR | 95% CI | P | HR | 95% CI | P | HR | 95% CI | P | |
| Mortality | 2.18 | 3.68 | 3.53–3.84 | 0.000 | 3.82 | 3.66–3.99 | 0.000 | 3.86 | 3.72–4.01 | 0.000 |
| Peripheral Nerve Injury | 0.09 | 1.20 | 0.99–1.47 | 0.067 | 1.08 | 0.88–1.33 | 0.472 | 1.10 | 0.75–1.30 | 0.928 |
| Coma | 0.13 | 3.42 | 2.88–4.05 | 0.000 | 3.42 | 2.88–405 | 0.000 | 3.45 | 2.97–4.02 | 0.000 |
| Myocardial Infarction | 0.23 | 2.07 | 1.85–2.31 | 0.000 | 2.28 | 2.04–2.55 | 0.000 | 2.22 | 1.98–2.49 | 0.000 |
| Graft Failure | 0.35 | 2.23 | 2.04–2.44 | 0.000 | 2.42 | 2.21–2.64 | 0.000 | 2.00 | 1.82–2.21 | 0.000 |
| Stroke/CVA | 0.37 | 2.40 | 2.19–2.62 | 0.000 | 2.36 | 2.16–2.58 | 0.000 | 2.36 | 2.16–2.57 | 0.000 |
| Pulmonary Embolism | 0.46 | 1.38 | 1.27–1.50 | 0.000 | 1.42 | 1.31–1.54 | 0.000 | 1.38 | 1.25–1.52 | 0.000 |
| Renal Insufficiency | 0.50 | 1.93 | 1.79–2.08 | 0.000 | 2.03 | 1.89–2.19 | 0.000 | 1.99 | 1.84–2.15 | 0.000 |
| Cardiac Arrest | 0.57 | 2.66 | 2.47–2.86 | 0.000 | 2.68 | 2.49–2.88 | 0.000 | 2.84 | 2.62–3.08 | 0.000 |
| Bleeding | 0.61 | 2.23 | 2.09–2.39 | 0.000 | 2.40 | 2.24–2.57 | 0.000 | 2.54 | 2.37–2.71 | 0.000 |
| Renal Failure | 0.64 | 2.46 | 2.30–2.63 | 0.000 | 2.72 | 2.54–2.91 | 0.000 | 2.72 | 2.54–2.90 | 0.000 |
The three approaches for unsupervised anomaly detection performed comparably, with both the k-nearest neighbor and cluster-based estimation methods having marginally higher hazard ratios than the one-class SVM.
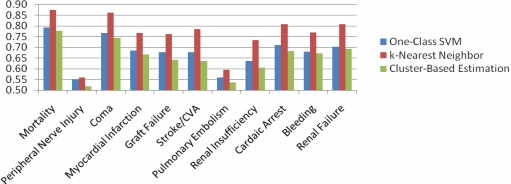
Figure 1 presents the AUROC for the different morbidity endpoints and mortality in the NSQIP data for each of the unsupervised anomaly detection approaches. In this case, the best results were obtained for all endpoints by the k-nearest neighbor approach. For eight of the 10 morbidity endpoints studied, this approach resulted in an AUROC higher than 0.7 (in three of these cases the AUROC was greater than 0.8). We obtained analogous results for the endpoint of mortality. While the one-class SVM performed worse than k-nearest neighbors in this patient population, it had a marginally higher AUROC than cluster-based estimation for all of the endpoints studied.
Figure 1:
AUROC of one-class SVM, k-nearest neighbor, and cluster-based estimation for morbidity and mortality outcomes in patients undergoing inpatient surgeries in the year 2008.
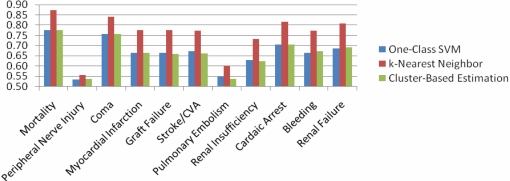
Figure 2 presents the average AUROC obtained across 2005–2008. In this case, our results mirrored our findings for 2008 alone, suggesting that the findings presented earlier generalized across the entire period.
Figure 2:
Average AUROC of one-class SVM, k-nearest neighbor, and cluster-based estimation for morbidity and mortality outcomes in patients undergoing inpatient surgeries in the years 2005–2008.
Consistent with our findings using Kaplan-Meier survival analysis, the AUROC showed a poor discrimination for the outcome of peripheral nerve injury. In addition, our results showed no clinically useful predictive ability for the outcome of pulmonary embolism.
Summary and Discussion
In this paper we presented and evaluated the hypothesis that patients who are anomalies in a population are at an increased risk of adverse outcomes. We described three different unsupervised anomaly detection methodologies to identify patients who are most different from other patients in the population, and investigated the utility of these approaches to preoperatively risk stratify patients undergoing inpatient surgical procedures.
Our results on over 100,000 patients show that unsupervised anomaly detection can help risk stratify patients without using extensive a priori information or needing large volumes of labeled historical data. In our study all three anomaly detection methodologies identified patients at an increased risk for morbidity and mortality in the 30-day period following inpatient surgery. We evaluated this effect both through Kaplan-Meier survival analysis and by measuring the AUROC. Intuitively, the AUROC can be considered as a measure of the discriminative ability of a risk variable, while Kaplan-Meier survival analysis measures the survival characteristics of patients in high (e.g., anomaly) and low (e.g., non-anomaly) groups. Both evaluation methods suggested that anomaly detection may be useful in risk stratifying patients for different adverse outcomes.
Two exceptions to this result were the morbidity outcomes of peripheral nerve injury and pulmonary embolism. For these outcomes, we did not find unsupervised anomaly detection to be useful in predicting adverse outcomes. We note that we only used baseline clinical characteristics in evaluating patients, and did not capture any information related to the management of these cases. This information may be particularly important in explaining outcomes such as peripheral nerve injury and pulmonary embolism. These two rare complications are highly preventable based on management countermeasures such as positioning and padding (for peripheral nerve injury) and Caprini scoring-based thrombo-prophylaxis regimens (for pulmonary embolism). It is therefore not surprising that these outcomes were difficult to predict from baseline characteristics alone. We also observe that our results for the one-class SVM may have been better for a different choice of the kernel parameter. Given our focus on unsupervised risk stratification, we did not perform any training to find an optimal choice for this parameter. As a result, our one-class SVM results were weaker than the results for methods such as k-nearest neighbors, which may be more robust to choices of parameters.
In the growing interest to improve patient outcomes, many such efforts are constrained by the immense costs related to data collection, verification, and maintenance. To make these efforts more widespread, solutions must be found that involve less extensive data collection and less intensive expert labeling. Outlier detection methodologies provide the opportunity to identify potential at-risk patients in a less data intensive environment, and could represent an important safety measure in the road ahead. We note that the results of this work can be improved further in future work, using ideas related to semi-supervised classification, and by exploring ways to combine the results of anomaly detection with the results of supervised approaches.
Footnotes
NSQIP Disclosure
The American College of Surgeons National Surgical Quality Improvement Program and the hospitals participating in the ACS NSQIP are the source of the data used herein; they have not verified and are not responsible for the statistical validity of the data analysis or the conclusions derived by the authors.
References
- 1.Khuri SF. The NSQIP: a new frontier in surgery. Surgery. 2005;138:837–843. doi: 10.1016/j.surg.2005.08.016. [DOI] [PubMed] [Google Scholar]
- 2.Schilling PL, Dimick JB, Birkmeyer JD. Prioritizing quality improvement in general surgery. J Am Coll Surg. 2008;207:698–704. doi: 10.1016/j.jamcollsurg.2008.06.138. [DOI] [PubMed] [Google Scholar]
- 3.Syed Z, Rubinfeld I. Unsupervised risk stratification in clinical datasets: identifying patients at risk of rare outcomes. International Conference of Machine Learning; Haifa, Israel. 2010. [Google Scholar]
- 4.Tarassenko L, Hayton P, Cerneaz N, Brady M. Novelty detection for the identification of masses in mammograms. Fourth International Conference on Artificial Neural Networks; Cambridge, UK. 1995. pp. 442–447. [Google Scholar]
- 5.Campbell C, Bennett KP. A linear programming approach to novelty detection. Advances in Neural Information Processing Systems. 2001:395–401. [Google Scholar]
- 6.Roberts S, Tarassenko L. A probabilistic resource allocating network for novelty detection. Neural Computation. 1994;6:270–284. [Google Scholar]
- 7.Laurikkala J, Juhola M, Kentala E. Informal identification of outliers in medical data. 5th International Workshop on Intelligent Data Analysis in Medicine and Pharmacology; Berlin, Germany. 2000. [Google Scholar]
- 8.Scholkopf B, Platt J, Shawe-Taylor J, Smola A, Williamson R. Estimating the support of a high dimensional distribution. Neural Computation. 2001;13:1443–1471. doi: 10.1162/089976601750264965. [DOI] [PubMed] [Google Scholar]
- 9.Vapnik VN. Statistical Learning Theory. New York: Wiley; 1998. [Google Scholar]
- 10.Eskin E, Arnold A, Prerau M, Portnoy L, Stolfo S. A geometric framework for unsupervised anomaly detection. Applications of Data Mining in Computer Security. 2002:77–200. [Google Scholar]
- 11.McCallum A, Nigam K, Ungar L. Efficient clustering of high dimensional data sets with application to reference matching. Knowledge Discovery and Data Mining; Boston, MA. 2000. pp. 169–178. [Google Scholar]