Abstract
Patient-specific data from electronic medical records (EMRs) is increasingly shared in a de-identified form to support research. However, EMRs are susceptible to noise, error, and variation, which can limit their utility for reuse. One way to enhance the utility of EMRs is to record the number of times diagnosis codes are assigned to a patient when this data is shared. This is, however, challenging because releasing such data may be leveraged to compromise patients’ identity. In this paper, we present an approach that, to the best of our knowledge, is the first that can prevent re-identification through repeated diagnosis codes. Our method transforms records to preserve privacy while retaining much of their utility. Experiments conducted using 2676 patients from the EMR system of the Vanderbilt University Medical Center verify that our method is able to retain an average of 95.4% of the diagnosis codes in a common data sharing scenario.
INTRODUCTION
The biomedical community is migrating toward the reuse of electronic medical records (EMRs) for large-scale population research, such as genome wide association studies (GWAS). Given the cost of such studies, the National Institutes of Health (NIH), among other organizations, have enacted policies that encourage researchers to share data.1,2 To facilitate this activity, various institutions have erected infrastructure to gather and disseminate records for reuse, such as the Database of Genotypes and Phenotypes (dbGaP).3 Data sharing must respect a patient’s right to privacy and, in support of this goal, many regulations require the data to be de-identified. For instance, the NIH policies invoke a standard, akin to the HIPAA Privacy Rule, which removes 18 identifiers (e.g., names and dates).4
Despite such measures, it has been demonstrated5 that EMR-derived records can be re-identified to named individuals when they retain standardized billing codes (e.g., ICD-9) that also reside in identified resources, such as discharge databases. This is a concern because the accompanying genomic data is often managed outside of the EMR6, such that linkage leads to the revelation of previously unknown sensitive data. Notably, computational methods have recently been designed to prevent this type of linkage attack in certain instances.7 However, these approaches assume a patient’s longitudinal record is summarized, such that it contains no more than one occurrence of a diagnosis. However, clinical informaticians have recognized that more detailed information is necessary, such that data from multiple visits8, with repeated codes, are utilized in EMR data mining activities. This type of profile is not addressed by existing privacy protection methods.
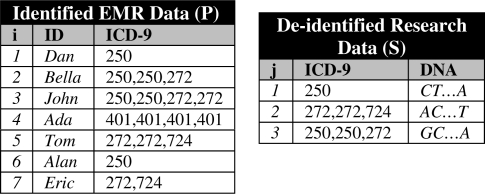
As an example of the problem studied in this work, consider the research dataset in the right of Figure 1. This was constructed by de-identifying a subset of the larger patient population’s clinical records, shown in the left of the figure, and assigning DNA data gathered in a research setting. If a data recipient knows that Tom was affiliated with the combination of codes “272, 272, 724”, they can associate Tom with his DNA sequence “AC…T”. This is because Tom’s record is the only one in the population with this code combination.
Figure 1.
left) Patient population dataset P and right) Research sample S.
In this paper, we present the first privacy-preserving approach to explicitly thwart this problem. We realize our approach in an automated algorithm, called Greedy Code Censoring (GCCens), which attempts to maximize the number of repeated codes that can be safely released without supporting linkage attacks. We illustrate the effectiveness of GCCens in retaining repeated comorbidities in a patient cohort from the Vanderbilt University Medical Center.9
BACKGROUND
The Need for Detailed EMR Data
EMR systems are not explicitly designed to support research and can accumulate misinformation that can limit reusability.10 One typical source of inaccuracies is administrative errors (e.g., when a patient is assigned ICD-9 code “250.00” for diabetes type I instead of “250.01” for diabetes type II). Another source is noise that arises from the healthcare delivery process (e.g., a specialist refines a diagnosis over time). While eliminating errors from EMR systems is challenging, researchers can still build accurate clinical models using EMR-driven data when this data contains repeated diagnosis codes (e.g., to identify phenotypes in the context of genetic association studies8). Typically, each time a patient is diagnosed with a condition or disease during a visit to the hospital, the patient’s record in the EMR system is updated by a clinician, which in turn, is transformed into an ICD-9 code for billing purposes. Hence, multiple visits to the hospital may result in the same ICD-9 code being replicated in a patient’s record, which can be applied to develop more accurate models of a patient’s status.
Data Privacy
An increasing number of investigations demonstrate that de-identified biomedical records5,11–14 are vulnerable to re-identification, often through publicly available resources. Several methods have been proposed to reduce re-identification risk by employing techniques that modify data prior to its release. Examples of such techniques are suppression, generalization, or randomization (see reference 15 for a survey). Suppression removes certain values from records (or entire records), whereas generalization replaces specific values with more general, but semantically consistent values. Randomization methods, on the other hand, add noise to the values.
In recent research5, it was shown that naïve application of suppression and generalization are both inadequate to preserve the privacy of EMRs containing diagnosis codes while retaining their utility in practice, but proposed no method to achieve this goal. A method to prevent the aforementioned linkage attack was proposed in 7, but was designed for data that contains no repeated diagnosis codes. In this paper, we propose a method to deal with the latter, more general case.
A Formal Model of the System
Before proceeding, we formalize the privacy problem considered in this paper. Let U be the set of diagnosis codes (henceforth referred to as codes) stored in an EMR system. The dataset P = {p1, …, pn} represents the medical records of the patient population. Each record pi is of the form 〈IDi, Di〉, where IDi is an identifier associated with a patient and Di = {d1, …, dk} is a set of codes for the patient (which are not necessarily distinct) derived from U. The table on the left of Figure 1 depicts a population that is comprised of seven records. The fifth record in this table has ID5 = Tom and D5 = {272, 272, 724}.
A second dataset S = {s1, …, sm} represents a sample of patient records to be shared. Each record sj is of the form 〈Dj, DNAj〉 and corresponds to a patient whose record is in the population. Dj is a set of codes derived from U and DNAj represents genomic sequence data. For instance, the record s1 in the table to the right of Figure 1 has D1 = {250}, DNA1 = {CT…A}, which was derived from record p6, that is Alan.
The linkage attack we consider assumes an attacker knows the identifying information and codes about a patient whose record is in the sample. This could occur through various routes. Consider, a data recipient may be an employee of the institution from which the data was derived, with access to the EMR system. Alternatively, the recipient may have knowledge about a neighbor or coworker.16 Or, in certain cases, a recipient may use public information; e.g., they may link de-identified hospital discharge summaries with identified resources, such as voter registration lists11,13.
METHODS
Materials
For this study, we worked with the de-identified version of StarChart, the EMR system of the Vanderbilt University Medical Center.6 We constructed P by using a set of 301,423 patients’ records that contain at least one of the following codes: “250” (diabetes mellitus), “272” (disorders of lipoid metabolism), “401” (essential hypertension), and “724” (back pain). We selected these codes because they appear frequently in the EMR system and are critical in defining a wide range of clinical phenotypes.
The research sample S contains 2676 patient records and was extracted for the purposes of a GWAS on native electrical conduction within the ventricles of the heart. The sample represents a “heart healthy” group with no prior heart disease, no heart conduction abnormalities, no electrolyte abnormalities, and no use of medications that can interfere with conduction. A record in S, on average, consists of 3.5, 2.3, 4,4, and 2 repeats of the codes “250”, “272”, “401”, and “724”, respectively.
A record in P, on average, consists of 2.2, 1.3, 2.5, and 0.9 repeats of the same codes. It was shown in previous research5 that this cohort is appropriate for studying privacy threats in samples derived from Vanderbilt’s EMR system.
Risk Measure
We measure the level of privacy protection afforded to the sample using the distinguishability measure.5 This measure is applied to determine how many records are susceptible to linkage based on shared diagnosis codes. Specifically, given a set of codes Dj in S, distinguishability (which we refer to as a function dis) is equal to the number of records in P that contain all the codes. For example, in Figure 1, dis(272, 724) = 2 because two records in P contain all of these codes (i.e., Tom and Eric). Distinguishability is the inverse of the probability of re-identifying a patient, such that we say a patient is uniquely distinguishable if his record has a distinguishability of 1.
Censoring Algorithm
GCCens is designed to limit the number of repeated codes that are released in patient records in a greedy manner. Greedy heuristics are commonly employed to anonymize data due to their ability to retain both privacy and utility relatively well.17 A notable strength of GCCens is that it significantly enhances data utility by employing a formal privacy model called k-map.13,18,19 This model states that each record in the sample can be associated with no less than k records in the population from which it was derived. In our setting, S satisfies k-map when, for each Dj in S, dis(Dj) ≥ k. This ensures that each record in S can be associated with no less than k records in P based on the released diagnosis codes, and implies that the probability of performing the linkage attack is no greater than 1/k.
The k-map model tends to offer “high” data utility, but assumes no knowledge of whether an individual’s record is contained in the released sample S.19 However, such knowledge is difficult (if not impossible) to be acquired by an attacker in the context of the data sharing we consider. This is because, typically, a random fraction of EMRs with identical codes are associated with DNA information and released.
More specifically, the GCCens algorithm accepts the following inputs: a sample S, a population P, a privacy parameter k and a set of censoring thresholds C. The algorithm outputs T, a version of S that is k-mapped to P. The parameter k expresses the minimum allowable number of records in P that can be mapped to a record of T based on diagnosis codes, while C is a set of thresholds (called caps), each of which corresponds to a distinct code in S and expresses the maximum allowable number of times a code can appear in a record of T. In effect, the caps act as an initial acceptable censor for the distribution of repeat counts. In this work, we follow standard assumptions13,20 in that we assume k and C are specified by data owners according to their expectations about an attacker’s knowledge. We also note that it is possible to specify C automatically by scanning S and recording the maximum number of occurrences of each code in all records.
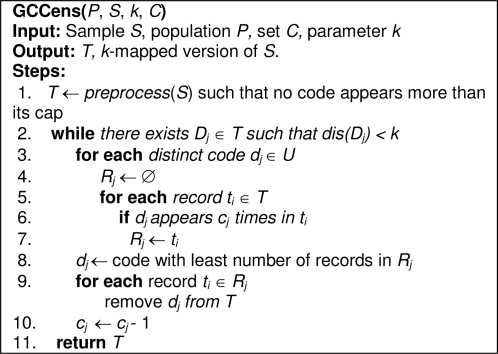
The pseudocode of GCCens is illustrated in Figure 2. In step 1, the algorithm invokes a helper function preprocess(), which iteratively censors diagnosis codes (i.e., removes one of their instances) from the dataset S until there is no code that appears more than its associated cap in S. The result of preprocess() is assigned to a dataset T. Then, in steps 2–10, GCCens iterates over the dataset T for as long as k-map is not satisfied. More specifically, in steps 3–7, the algorithm computes the number of code instances that need to be censored for each distinct code dj in U. This is achieved by iterating over all records in T (step 3), counting the number of records that harbor a diagnosis code dj that appears cj times in T and assigning these records to a set Rj (steps 5–7). Then, in steps 8–9, the algorithm determines the code dj that requires the least amount of censoring and removes it from all the records in Rj. To minimize the number of codes that need to be modified, we remove one instance of dj per iteration. Subsequently, the cap cj for the censored code is decremented by 1 (step 10). Finally, GCCens releases a sample that satisfies k-map in step 11.
Figure 2.
Pseudocode for the GCCens algorithm.
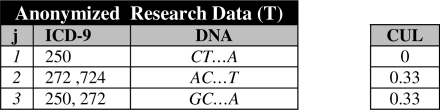
As an example, let’s walk through the application of GCCens to the records in Figure 1. We assume k = 2 and the censoring caps for the codes {250, 272, 401, 724} are C = {2, 2, 0, 1}. First, T is set equivalent to S because no code appears more than its cap. Next, GCCens finds that 2-map is not satisfied because dis(D3 = {250, 250, 272}) = 1. So, GCCens censors one occurrence of “250” from the third record of T (i.e., this code was selected because the cap value is 2 and only one record in T has 2 instances of the code). After censoring, the cap value for “250” is decremented by 1, so C = {1, 2, 0, 1}. At this point, GCCens finds k-map is still not satisfied because dis(D2 = {272, 272, 724}) = 1. Thus, one occurrence of “272” is censored from the second record of T. Finally, T satisfies 2-map and the algorithm terminates. The resulting solution is depicted in Figure 3.
Figure 3.
The resulting research sample from output by GCCens.
Data Utility Measure
When diagnosis code repeats are censored, there is a decrease in the utility of the dataset. To measure the utility loss, we introduce a measure called Censoring Utility Loss (CUL). This is defined as the number of censored codes in a record s divided by the total number of codes in s. As an example, assume that we remove one instance of “272” from the second record in S shown on the right table of Figure 1. In this case, CUL equals 1/3 because there were three codes in this record, one of which is censored. We note that the greedy heuristic in GCCens is designed to minimize the sum of CUL values in each iteration by choosing to censor the code that incurs the minimum utility loss (see step 8 in Figure 2).
RESULTS
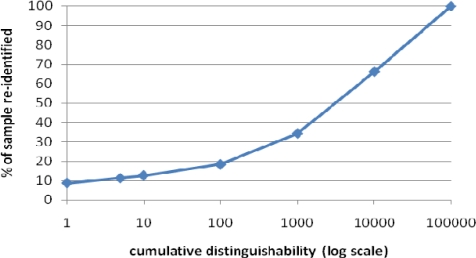
First, Figure 4 summarizes the risk of associating a patient’s record from the de-identified sample to their corresponding record in the patient population. This figure is a cumulative distribution and depicts the percent of patients in the sample (y-axis) that have a distinguishability score of a particular value or less (x-axis) with respect to the population from which they were derived. As can be seen, more than 9% of the patients contained in the sample would be uniquely identifiable if the original data were disclosed. This confirms that a linkage attack is feasible in practice and the need for developing a formal protection method.
Figure 4.
Distinguishability of the original patient records in the sample. A distinguishability of 1 means that a patient is uniquely identifiable.
Next, to evaluate the effectiveness of GCCens in preserving utility, we report CUL scores when it is applied with all caps set to 3 (i.e., C = {3,3,3,3}) and various k values between 2 and 25. Table 1 reports the mean, standard deviation, median, and skewness (this is a standard measure of the asymmetry of the distribution21) of the distribution of CUL values for all records in the sample dataset. As expected, as we increase k we find an increase in the mean of the distribution of CUL. This is because GCCens needs to censor a larger number of codes to meet a stricter privacy requirement. However, it is notable that GCCens retained 95.4% of the codes on average when k = 5 as is often applied in practice (i.e., the mean of the distribution of CUL was 0.046).7 We note that while 4.6% of the codes in a record were censored on average, GCCens modified 16% of the records in S. We also observed a positive skew in the distribution of CUL for all tested values of k, which implies that the number of censored codes is closer to 0 for most patient records.
Table 1.
Statistics on the distribution of CUL when GCCens was applied with all caps set to 3.
| k | Mean | Std. Dev. | Median | Skewness |
|---|---|---|---|---|
| 5 | 0.046 | 0.123 | 0 | 3.016 |
| 10 | 0.046 | 0.123 | 0 | 3.016 |
| 25 | 0.091 | 0.156 | 0 | 1.501 |
Finally, we recognize that not all data recipients will feel comfortable working with EMR data this capped to varying degrees. Thus, we evaluated the impact of forcing all values in C to be equivalent. For this set of experiments, we fixed k to 5 and varied the cap between 3 and 10. The results are summarized in Table 2. Notice that GCCens performed a greater amount of censoring when larger cap values are supplied. This is expected because large cap values permit more information to be released, which makes it more difficult to generate a sufficient privacy solution.5,15,20 However, GCCens managed to retain a reasonably large percentage of codes in all tested cases. In particular, 92% of codes were retained when the universal cap was set to 4 (i.e., the mean of the CUL distribution was 0.08). We believe this result is promising because the dataset derived by GCCens in this experiment was deemed to be useful for comorbidity analysis. Moreover, when releasing 5 diagnosis codes GCCens retained on average 88.1% of the codes.
Table 2.
Distribution statistics of CUL when GCCens was applied with k = 5 and a universal cap.
| Cap | Mean | Std. Dev. | Median | Skewness |
|---|---|---|---|---|
| 3 | 0.046 | 0.123 | 0 | 3.016 |
| 4 | 0.080 | 0.152 | 0 | 2.042 |
| 5 | 0.119 | 0.183 | 0 | 1.383 |
| 6 | 0.141 | 0.209 | 0 | 1.131 |
| 7 | 0.156 | 0.229 | 0 | 1.050 |
| 8 | 0.191 | 0.270 | 0 | 0.952 |
| 9 | 0.197 | 0.279 | 0 | 0.939 |
| 10 | 0.213 | 0.282 | 0 | 0.898 |
DISCUSSION
In this paper, we demonstrated the feasibility of a linkage attack based on repeated diagnoses derived from real patient-specific clinical data and developed an algorithm to provide formal computational guarantees against this attack. Our experiments verify that the proposed approach permits privacy-preserving patient record dissemination while retaining much of the information of the original records.
We believe this work is an important step towards increasing the type of information that can be made available to researchers without compromising patients’ privacy rights. This is partly because the approach we propose can be directly utilized by researchers when depositing data with diagnosis codes to repositories. However, our approach is limited in certain aspects that we wish to highlight to initiate further studies.
First, as is true for all privacy-preserving approaches, our approach makes certain assumptions about the maximum amount of knowledge an attacker may possess and the semantics of published data. As such, it does not offer privacy protection guarantees against attackers who are able to exploit additional information (e.g., the time between a patient’s hospital visits) that may be published together with the type of data we studied in this paper. Information such as the relative time between diagnoses may be beneficial to assess chronicity, disease evolution, and downstream comorbidities. For example, patients with rheumatoid arthritis may later need joint replacements, and have an increased incidence of cardiovascular disease. We are currently working towards extending our approach to allow this type of information to be released in a privacy-preserving way.
Second, our approach suppresses diagnosis codes from the released dataset, which may incur more information loss compared to alternative data modification strategies that have been successfully applied on biomedical data, such as generalization.7 As part of our future work, we intend to examine whether these strategies can be used alone or in combination with our approach to further enhance data utility.
Acknowledgments
This research was funded by NIH grants R01LM009989 and U01HG004603.
References
- 1.National Institutes of Health Final NIH statement on sharing data. NOT-OD-03-032. 2003.
- 2.National Institutes of Health . 2007. Policy for sharing of data obtained in NIH supported or conducted genome-wide association studies. NOT-OD-07-088. [Google Scholar]
- 3.Mailman M, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat Genet. 2007;39:1181–6. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Dept of Health and Human Services Standards for protection of electronic health information; Final Rule. Federal Register. 2003 45 CFR: Pt. 164. [Google Scholar]
- 5.Loukides G, Denny J, Malin B. The disclosure of diagnosis codes can breach research participants privacy. J Am Med Inform Assoc. 2010;17:322–7. doi: 10.1136/jamia.2009.002725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roden DM, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin Pharmacol Ther. 2008;84:362–9. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Loukides G, Gkoulalas-Divanis A, Malin B. Anonymization of electronic medical records for validating genome-wide association studies. Proc Nat Acad Sci. 2010;107:7898–903. doi: 10.1073/pnas.0911686107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ritchie MD, et al. Robust replication of genotype-phenotype associations across multiple diseases in an electronic medical record. Am J Hum Genet. 2010;86:560–72. doi: 10.1016/j.ajhg.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Electronic Medical Records & Genomics Network URL: http://www.gwas.net.
- 10.Singh H, et al. Identifying diagnostic errors in primary care using an electronic screening algorithm. Arch Intern Med. 2007;167:302–8. doi: 10.1001/archinte.167.3.302. [DOI] [PubMed] [Google Scholar]
- 11.Benitez K, Malin B. Evaluating re-identification risks with respect to the HIPAA privacy rule. J Am Med Inform Assoc. 2010;17:169–77. doi: 10.1136/jamia.2009.000026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.El Emam K, et al. Evaluating common de-identification heuristics for personal health information. J Med Internet Res. 2006;8:e28. doi: 10.2196/jmir.8.4.e28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sweeney L. k-anonymity: a model for protecting privacy. International Journal of Uncertainty, Fuzziness, and Knowledge-based Systems. 2002;10:557–70. [Google Scholar]
- 14.Malin B. An evaluation of the current state of genomic data privacy protection technology and a roadmap for the future. J Am Med Inform Assoc. 2005;12:28–34. doi: 10.1197/jamia.M1603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen B, Kifer D, Lefevre K, Machanavajjhala A. Privacy-preserving data publishing. Foundations and Trends in Database Systems. 2009;2:1–167. [Google Scholar]
- 16.Machanavajjhala A, et al. l-diversity: privacy beyond k-anonymity. Proc IEEE International Conference on Data Engineering. 2006:24. [Google Scholar]
- 17.Loukides G, Shao J. Clustering-Based k-Anonymisation algorithms. In the Proc of the 18th International Conference on Database and Expert Systems Applications (DEXA) 2007 [Google Scholar]
- 18.Malin B. k-unlinkability: a privacy protection model for distributed data. Data and Knowledge Engineering. 2008;64:294–311. [Google Scholar]
- 19.Sweeney L. Laboratory for International Data Privacy. Pittsburgh, PA: 2000. Computational Data Privacy Protection LIDAP-WP5. Carnegie Mellon University. [Google Scholar]
- 20.Xu Y, Wang K, Fu AWC, Yu PS. Anonymizing transaction databases for publication. Proc ACM SIGKDD Conference. 2008:767–75. [Google Scholar]
- 21.Sullivan M. Fundamentals of statistics. Prentice Hall; Upper Saddle River, NJ: 2010. [Google Scholar]