Abstract
Text simplification is a challenging NLP task and it is particularly important in the health domain as most health information requires higher reading skills than an average consumer has. This low readability of health content is largely due to the presence of unfamiliar medical terms/concepts and certain syntactic characteristics, such as excessively complex sentences. In this paper, we discuss a simplification tool that was developed to simplify health information. The tool addresses semantic difficulty by substituting difficult terms with easier synonyms or through the use of hierarchically and/or semantically related terms. The tool also simplifies long sentences by splitting them into shorter grammatical sentences. We used the tool to simplify electronic medical records and journal articles and results show that the tool simplifies both document types though by different degrees. A cloze test on the electronic medical records showed a statistically significant improvement in the cloze score from 35.8% to 43.6%.
Introduction
Text simplification is a challenging natural language processing (NLP) task. Its application to the health domain is of special importance as the majority of health information, including those articles targeted at consumers, require reading skills that an average US adult does not have1. This significantly limits the impact of the available health information on improving health outcomes.
Tools to automate the simplification of health content can be instrumental in making health information more accessible to consumers. There has been relatively limited prior research on such tools2. One of the few simplification tools that address this issue is a system developed by Elhadad3 that identifies difficult terms and retrieves definitions for these terms using the Google search engine. Retrieved definitions are provided as links to the terms. The system was found to improve reader’s comprehension by an average 1.5 points on a 5 point scale.
In a 2007 paper, we described an ad hoc method for simplifying health content4. The method addressed the semantic difficulty of health content that results from the use of terms unfamiliar to an average reader. The method identified difficult terms in the text and tried to simplify them either by replacing them with easier synonyms or by explaining them using easier related terms. The study reported useful and correct simplifications in 68% of identified difficult terms.
The simplification tool discussed here extends this earlier tool by using a larger set of relationship types that were identified through a more systematic review of human-generated explanations. Additionally it incorporates a module to address the syntactic difficulty of health texts. Though we recognize that syntactic difficulty can arise due to a number of reasons, in this version of the tool we only focus on identifying and simplifying complex and compound sentences, i.e. sentences with at least one dependent clause and one independent clause.
To validate the system, we used two types of health documents – electronic medical records and biomedical journal articles. Through the increasing availability of personal health records and services like PubMed, the general public now has access to both these types of difficult documents. However, these texts require literacy and numeracy skills much higher than that of an average reader. The readability of electronic medical records is low due to extensive use of medical terms and abbreviations, short ungrammatical sentences and very little cohesion. Journal articles are generally more cohesive but they use difficult terms and excessively long sentences with complicated syntactic structures which are hard to follow. In the following sections, we describe the different modules of the system, report results of the tests and discuss their implications.
Background
Identification of difficult terms is the primary step in text simplification. In a previous study5, we have proposed a frequency-based technique to estimate the difficulty of terms. This method is based on the observation that terms that occur more frequently in lay reader targeted biomedical sources, such as Reuters News or MedlinePlus queries, tend to be easier. We have also proposed a contextual network approach that estimates a term’s difficulty based on its usage contexts6. Both methods have been validated using data from actual consumer surveys. A combination of these two techniques can be used to obtain a single score, familiarity score, in the range of 0 (very hard) and 1 (very easy) as an estimate of a lay reader’s familiarity with a given term. All terms with a familiarity score less than a user-defined threshold can be considered to be difficult and in need of simplification. The simplification tool described by Zeng-Treitler et al.4 uses the familiarity score to identify difficult terms. Terms with a familiarity score below a specified threshold were simplified through one of the following:
Synonym replacement
A simple approach to simplify difficult terms is to replace them with their easier synonyms. Prior studies have shown that there are significant differences between a patient’s and a healthcare professional’s mental model of the medical domain7,8 and they prefer different terms to describe the same concept. The Open Access and Collaborative Consumer Health Vocabulary (OAC CHV) Initiative9 is an on-going effort to curate consumer-friendly alternatives to difficult medical terms. Difficult terms identified for simplification using their familiarity score, can have OAC CHV-preferred synonyms that satisfy the familiarity score threshold criterion. If such synonyms exist, the difficult terms were replaced with their OAC CHV-preferred synonyms.
Explanation Generation
For difficult terms that have no OAC CHV-preferred synonyms, explanations were generated in an ad hoc manner. An explanation is defined as a short phrase that makes the term more understandable and is different from the term’s definition which is focused on describing the precise and complete semantics. As such, a definition can use terms more difficult than the original term, but an explanation cannot; a definition may also need to be lengthy but an explanation should be brief. For example, a definition (MeSH) for hemochromatosis can be ‘a disorder due to the deposition of hemosiderin in the parenchymal cells, causing tissue damage and dysfunction of the liver, pancreas, heart, and pituitary’ and an explanation (MedlinePlus) can be ‘an inherited disease in which too much iron builds up in the body’.
To automatically generate explanations, the tool: (a) generates a pool of terms related to the original term; (b) selects the easiest related term using the familiarity score; and, (c) uses a short phrase to describe the relationship between the difficult term and the selected related term. To generate a pool of related terms, synonymous (SY) and hierarchical relationships (PAR, CHD) defined in the Unified Medical Language System (UMLS) Metathesaurus were used. For example, UMLS defines disordered taste as a synonym of dysgeusia and congenital abnormality as a parent of pulmonary atresia.
Related terms that have an OAC CHV alternative were replaced with the corresponding OAC CHV term. The familiarity score of the related term was calculated and the easiest related term that satisfies the familiarity score threshold criterion was selected. If the selected term is a synonym it was used to replace the original term. If the selected term is a parent of the original term, the replacement was of the form <difficult_term> (a type of <parent>). Similarly, if it is a child, the replacement was of the form <difficult_term> (e.g. <child>). Pulmonary atresia (a type of birth defect) and oropharyngeal (e.g. mouth) are examples of generated explanations.
A human expert review found 32% of the explanations to be either not useful or incorrect. One major source of these errors was the use of non-applicable hierarchical relations (for example, ‘tobacco abuse’ as ‘a type of psychiatric problem’). This led us to believe that supplementing the hierarchical relations with other relations might reduce these errors and improve the simplification.
Methods
Building on the functionality described in the previous section, we extended the tool to use additional relationship types for explanation generation and added a new syntactic component.
Semantic Simplification
In a related ongoing study, we manually analyzed explanations contained in a set of 150 human created diabetes-related documents and identified key relations used to explain five common semantic groups10 of health concepts (Table 1). We found that the relationship types between the difficult term and the easier explanation terms are very often dependent on the semantic type of the difficult term.
Table 1.
Connectors used in explanations generated using semantic type information
| Original Term Sem. Group | Connector | Explanation Term Sem. Group |
|---|---|---|
| Disease name | a condition affecting | Anatomical Structure |
| Anatomical Structure | a part of | Anatomical Structure |
| Device | a device/instrument used in | Procedure |
| Procedure | a procedure performed on | Anatomical Structure |
| Medication | can have a tradename of | Medication |
For instance, it was found to be common to explain a disease/condition with common symptoms observed or the specific body parts affected. Hence, we modified the explanation generation algorithm to take into consideration the semantic type of the difficult term and to generate semantically influenced explanations. For example, the condition Endometrial adenocarcinoma is explained as Endometrial adenocarcinoma (a condition affecting female genitalia) and Humerus as Humerus (a part of arm).
For difficult terms that have both hierarchical and semantic explanations, the semantic explanation is appended to the hierarchical explanation. For example, polynephritis is explained as a type of infection affecting renal pelvis or urinary tract.
Syntactic Simplification
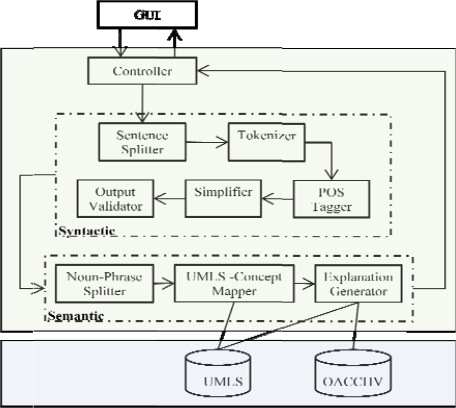
The syntactic simplification is performed at the sentence level. Sentences longer than 10 words are assumed to require syntactic simplification and are processed through a series of modules (discussed below). At the end of this simplification the original sentence can be retained unchanged or replaced by two or more, shorter, hence presumably simpler, sentences. The modules include (also see Figure 1):
Figure 1.
Architecture diagram with system modules
Part of Speech Tagger
A sentence that meets the length criterion is tokenized and each token is annotated with appropriate part-of-speech (POS) tag, as required by the simplifier module in the next step. We used open source software from OpenNLP11 to perform these tasks.
Grammar Simplifier
This module breaks down the long sentence into two or more shorter sentences and is based on the simplifier proposed by Siddharthan12. The module works by identifying POS patterns and applies a set of transformational rules to produce shorter sentences. For example this module splits the sentence “Although the subjects with high CACSs may be at higher risk of a first event, further follow-up data are needed before EBT screening can be recommended for type 1 diabetic patients.” into “The subjects with high CACSs may be at higher risk of a first event.” and “But further follow-up data are needed before EBT screening can be recommended for type 1 diabetic patients.”
Output Validator
To guard against ungrammatical and fragmented simplifications, the output validator module checks each output sentence pr roduced by the above module against the following conditions:
sentence too short (word count <7) – during testing we observed that such sentences are often fragmented texts and do not improve readability;
sentence has more than one null link - link grammars13, in order to discover the linkage structure of a given sentence, identify token(s) in the sentence that hinder a grammatical interpretation of the sentence. These tokens are represented as null links and the number of null links is inversely related to the soundness of a sentence’s syntax (zero links is ideal; the fewer, the better).
low OpenNLP score – the score is the log of the product of the probability associated with all the decisions that formed the constituent parse. If this score is less than -6, the sentence is very unlikely to be a reasonable English sentence. The cutoff of -6 was determined empirically.
If any of the output sentences satisfies one of the above conditions, the simplification is rejected and the original sentence is retained. Otherwise, the original sentence is replaced by the simplifier’s output. For the example given above, both output sentences satisfy all three validation conditions and hence the original sentence is replaced.
Validation
The tool described here can potentially be used to simplify any type of health text. To validate the system, we chose to test the tool on two sets of health related documents: (i) electronic medical recordsa (n=40); and (ii) biomedical journal articlesb (n=40). For these two document sets, we assessed the readability of the original and simplified texts using the popular readability formulae, Flesch Kincaid Grade Level (FKGL) and Simple of Measure of Gobbledygook (SMOG). Since FKGL and SMOG measure limited readability characteristics, we also report the following measures:
Semantic difficulty: the average familiarity score (defined in the Methods section) of all terms in the document.
Cohesion: the term overlap ratio, the ratio of the number of stemmed14 term overlaps between adjacent sentences to the total number of distinct terms in the text, is a proxy for cohesion. A high overlap ratio is assumed to indicate good cohesion.
In addition to these 80 documents, we performed a detailed analysis of the simplification of nine separate electronic medical records that were used to test the previous version of this tool4. We performed a cloze test15 on eight of the nine documents using four reviewers. In this test every fifth word of the first 250 words of the document was replaced with a blank and the reviewers were asked to guess the missing word. A higher percentage of correct guesses is considered to indicate simpler text. We assigned each reviewer eight documents (four before simplification and four after simplification; no reviewer is assigned both versions of the same document) and each document was reviewed by two reviewers. All reviewers have at least an undergraduate degree, have a non-clinical background and three are native speakers of English.
The average accuracy percentage for each document was computed and used to build a linear regression model with the cloze score as the dependent variable and the reviewer, document and document type (original/simplified) as categorical independent variables. A significant p-value for the regression was used as a test of the effectiveness of the simplification process.
Results
Table 2 shows the average readability measures for journal articles before and after simplification. The results show that the readability improved by all four measures. FKGL and SMOG reading grade levels dropped marginally. Both these formulae use average sentence length and syllable count to calculate grade levels. The average number of syllables/word was found to have decreased (from 1.83 to 1.80) but the average number of words/sentences increased slightly (from 25.23 to 25.52) owing to the addition of explanations. Both the familiarity score and cohesion improved.
Table 2.
Change in readability of journal articles
| Original | Simplified | |||
|---|---|---|---|---|
| Mean | SDc | Mean | SD | |
| FKGL | 15.83 | 1.44 | 15.58 | 1.43 |
| SMOG | 17.27 | 1.10 | 17.08 | 1.08 |
| Familiarity Score | 0.75 | 0.04 | 0.77 | 0.04 |
| Cohesion | 0.66 | 0.18 | 0.71 | 0.20 |
Table 3 shows the corresponding results for the electronic medical records. While the improvements in familiarity score and cohesion are consistent with those seen in journal articles (though they have relatively low cohesion), the grade levels increased. Additional analysis showed that the average words/sentence increased quite significantly from 10.62 to 13.30 whereas average/syllable word remained unchanged (1.67 to 1.66). Although the explanations added to electronic medical records are similar to those added to journal articles, these additions do not add as much to the long journal sentences as they do to the sentences in electronic medical records, which tend to be very short.
Table 3.
Change in readability of electronic medical records
| Original | Simplified | |||
|---|---|---|---|---|
| Mean | SD | Mean | SD | |
| FKGL | 8.31 | 1.78 | 9.23 | 2.12 |
| SMOG | 11.31 | 1.45 | 12.27 | 1.87 |
| Familiarity Score | 0.73 | 0.04 | 0.76 | 0.03 |
| Cohesion | 0.09 | 0.07 | 0.10 | 0.06 |
A paired t-test found the differences in these four measures for both document sets to be statistically significant (p <0.01).
The average cloze score for the reports before simplification was found to be 35.8% (min=17.2%, max = 54%; median = 36%). After simplification, the score improved to 43.6% (min = 36.7%, max= 57.4%, median = 40.8%). For the regression model built using the reviewer, document and document type (original/simplified) as categorical independent variables and the cloze score as dependent variable, the p-value for the regression as a whole was found to be significant (p < 0.001).
Discussion
We have discussed the development of a tool that simplifies health content using available biomedical sources. We tested the tool using four measures of readability on two types of health documents and a cloze test on a smaller sample. The tool simplified journal articles by all four calculated measures. The increase in grade level of electronic medical records due to the simplification is not a reason for concern as these readability formulae are generally recognized to be unsuitable for this type of documents; the clinical notes are not of 8th or 9th grade levels as the formulae incorrectly estimate. In medical records, vocabulary difficulty is of greater concern and the results show that both vocabulary and cohesion have been improved by the tool.
Although all the improvements are statistically significant, the magnitude of improvements is rather small. We believe this is due to the relatively fewer instances of syntactic simplification compared to the number of added explanations, which increased the average sentence length and hence the grade levels. Adding explanations as separate sentences can reduce the overall grade levels but this may not be appropriate in certain contexts and needs to be explored further.
It is important to note that the measures reported here are only surrogates to the lay users’ understanding and direct user testing is a more reliable means to test readability. The limited user testing of our tool showed significant improvement in cloze score although the score is not yet in the desirable score range (a score of 50%–60% is considered to represent reasonable readability). More comprehensive user testing with a larger document set is necessary for a more reliable validation of the tool.
The results also show that the new changes helped improve the performance of the tool over that of the earlier version4. While for the previous version of the tool the improvement in the cloze score was not statistically significant, the current version yields statistically significant results. A comparison of the improvements is not appropriate as we used different numbers of users and a different review scheme.
Related Issues
The estimate of cohesion used here is rudimentary and we are aware that the Coh-Metrix project has a more in-depth analysis of cohesion and other related factors. We have explored using some of these measures but we have been advised by our Coh-Metrix contact that there is neither a single cohesive measure of text readability (like FKGL/SMOG) that aggregates the different measures nor a method to establish corpus-independent criteria to choose a few key measures to report.
The explanation generation method employed here, uses key phrases as connectors between a difficult term and the related easy (easier) term. Though we have identified appropriate connectors for some of the more commonly occurring semantic types, this work needs to be extended to include other semantic types. A related challenge is to identify sources that describe the relationships between concepts in the level of detail required to generate meaningful and useful explanations.
Acknowledgments
This work is supported by grants from the National Institute of Diabetes and Digestive and Kidney Diseases (R01 DK 075837) and the National Institute of Health (R01 LM07222). We thank Advaith Siddharthan for providing a version of the syntactic simplifier.
Footnotes
Source: Clinical office notes and discharge summaries from Brigham and Women’s Hospital, Boston, MA
Source: DiabetesCare, Annals of Internal Medicine, Circulation, Journal of Clinical Endocrinology and Metabolism; British Medical Journal
Standard Deviation
References
- 1.Nielsen-Bohlman L, Panzer AM, Kindig DA. Health literacy: A prescription to end confusion. Natl Academy Pr. 2004 [PubMed] [Google Scholar]
- 2.Keselman A, Logan R, Smith CA, Leroy G, Zeng-Treitler Q. Developing informatics tools and strategies for consumer-centered health communication. J Am Med Inform Assoc. 2008 Jul-Aug;15(4):473–83. doi: 10.1197/jamia.M2744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Elhadad N. Comprehending technical texts: predicting and defining unfamiliar terms. AMIA Annual Symposium proceedings / AMIA Symposium. 2006:239–43. [PMC free article] [PubMed] [Google Scholar]
- 4.Zeng-Treitler Q, Goryachev S, Kim H, Keselman A, Rosendale D. Making texts in electronic health records comprehensible to consumers: a prototype translator. AMIA Annual Symposium proceedings / AMIA Symposium. 2007:846–50. [PMC free article] [PubMed] [Google Scholar]
- 5.Zeng Q, Kim E, Crowell J, Tse T. A text corpora-based estimation of the familiarity of health terminology. Vol. 3745. Springer; 2005. p. 184. [Google Scholar]
- 6.Zeng-Treitler Q, Goryachev S, Tse T, Keselman A, Boxwala A. Estimating consumer familiarity with health terminology: a context-based approach. J Am Med Inform Assoc. 2008 May-Jun;15(3):349–56. doi: 10.1197/jamia.M2592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Slaughter L, Keselman A, Kushniruk A, Patel VL. A framework for capturing the interactions between laypersons’ understanding of disease, information gathering behaviors, and actions taken during an epidemic. Journal of biomedical informatics. 2005 Aug;38(4):298–313. doi: 10.1016/j.jbi.2004.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rotegard AK, Slaughter L, Ruland CM. Mapping nurses’ natural language to oncology patients’ symptom expressions. 2006;122:987. [PubMed] [Google Scholar]
- 9.Zeng QT, Tse T, Crowell J, Divita G, Roth L, Browne AC. Identifying consumer-friendly display (CFD) names for health concepts. AMIA Annual Symposium proceedings / AMIA Symposium. 2005:859–63. [PMC free article] [PubMed] [Google Scholar]
- 10.Bodenreider O, McCray AT. Exploring semantic groups through visual approaches. Journal of biomedical informatics. 2003 Dec;36(6):414–32. doi: 10.1016/j.jbi.2003.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Morton T, Kottmann J, Baldridge J, Bierner G. 2005. OpenNLP: A Java-based NLP Toolkit. ;
- 12.Siddharthan A. Syntactic simplification and text cohesion. Vol. 4. Springer; 2006. pp. 77–109. [Google Scholar]
- 13.Grinberg D, Lafferty J, Sleator D. A robust parsing algorithm for link grammars. 1995.
- 14.Porter MF. An algorithm for suffix stripping. Program. 1980;14:130–7. [Google Scholar]
- 15.Taylor WL. Recent developments in the use of cloze procedure. Journalism Quarterly. 1956;33:42–8. [Google Scholar]