Abstract
Physicians have access to patient notes in volumes far greater than what is practical to read within the context of a standard clinical scenario. As a preliminary step toward being able to provide a longitudinal summary of patient history, methods are examined for the automated extraction of relevant patient problems from existing clinical notes. We explore a grounded approach to identifying important patient problems from patient history. Methods build on existing NLP and text-summarization methodologies and leverage features observed in a relevant corpus.
Introduction
Advances in medical informatics have led to the capture of high volumes of medical data in coded form, but a significant percent of crucial patient information remains embedded in clinical narratives. Physicians regularly have access to a far greater collection of notes than may readily be reviewed before or during the patient encounter. We have proposed1 that an automatically generated longitudinal patient summary would go far to helping physicians understand key aspects of patient history. The patient’s clinical problems are likely to be at the core of the summary. This is not surprising; since Larry Weed’s problem-oriented medical record2, clinicians and researchers have espoused the logic of organizing medical documentation around the patient’s clinical problems. Furthermore, the Institute of Medicine and JCAHO place great importance on the relevance of the clinical problem list. If it is accepted that the first step in summarizing patient information is to extract a problem list from the notes, the challenge remains to identify the subset of problems, which are actually relevant for the patient at present day, among the large number of all the problems mentioned in the notes. This study examines methods for best selecting such relevant problems from the notes.
Advances in Natural Language Processing (NLP) have facilitated the parsing of text-based information into structured output, however little work has been carried out in the application of high-level NLP applications, such as summarization, to this genre of texts. In this study, we investigate how to identify the problems that are mentioned in the patient notes and that are relevant to a physician seeing a new patient at present day. Given a corpus of patient records, each consisting of a set of NLP-parsed notes, we investigate corpus-based, bottom-up approaches to the selection of clinical problems for a patient summary. In particular, we explore a novel set of features for identifying relevant problems and cast our task as a classification model. The resulting model is robust and generalizable, applicable to any clinical specialty.
Related Work
Past research on problem list extraction/generation has investigated identifying active patient problems from clinical notes3,4, though much of it relies on a fixed set of problems, determined in a top-down fashion. As shown by Rassinoux et al.5, there is great power in the combination of medical knowledge stored in clinical terminologies with information mined from clinical narrative. This research leverages knowledge in the UMLS6, including SNOMED7 and MEDCIN in particular. We augment the knowledge-based approach employed in previous work on problem list extraction with a machine learning approach, to learn rules for a context-aware problem selection tool expected to be generalizable to other specialties.
Problem list extraction methods have drawn on work in the broader field of clinical NLP, which has been approached in numerous research projects including medSYNDIKATE8, MENELAS9 and the NLM’s MetaMap10 with modifications such as NegEx11. In this work, we rely on MedLEE12, a context-aware clinical natural language processor developed at Columbia, for the initial pre-processing of our dataset. MedLEE results are used to identify UMLS codes for identifiable clinical concepts and semantic indicators for negation in the source text.
Application of machine learning techniques to medical text summarization has focused primarily on associating relevant external literature to a current patient13,14.
Methods
This research approaches the selection of clinical problems relevant to a patient summary as a classification problem. The general architecture of the problem selection is as follows: concepts and their associated semantic types are identified in the notes of a patient. For every problem mentioned in the notes, a binary classifier determines whether the problem should be included in the patient summary. The classifier is trained on a collection of patient notes and their corresponding problem lists. We describe next the data used to train and test the classifier, the features we propose for the task, the learning models we experimented with, and our evaluation methodology.
Data Collection:
In order to train a classifier to predict the relevance of a problem for a clinical summary, we experimented with two datasets.
Renal Note Corpus:
As in-depth patient summaries are rarely authored in clinical medicine, the model was trained on the closest surrogate to a patient summary available in standard clinical notes: the patient’s last available past medical history (PMH). While the PMH is by no means a comprehensive, longitudinal patient history, it is the closest thing available on a large scale. The PMH may not always contain the immediate chief complaint, however for a summary we were interested in problems with lasting relevance, so this is, if anything, a benefit. To focus on problems with lasting relevance, we required that the last PMH have three or more problems listed and that the preceding two (or more if occurring within a week) notes were omitted from the patient corpus in order to focus on problems with lasting value.
The corpus analyzed under this model included 1618 patients having visited the NewYork-Presbyterian outpatient renal service from November 2007 through September 2009, with an average 14.3 notes per subject. This is referred to as the Renal Corpus.
Expert Summaries:
Nephrologists were asked to review existing notes for four renal patients and compose free-text summaries containing information relevant to a physician trying to acquaint him/herself with each patient’s medical history. The source notes used in summarizing these four patients did overlap with the Renal Corpus but were not a subset as notes were assessed from 2000 through September 2009. For these four patients the corpus contained an average of 49.5 notes per patient. These expert summaries served as the gold standard against which we evaluated the relevance of problems selected for the summary. Problems occurring in expert summaries not observed in the notes were ignored.
Problem Identification:
The unit of processing for the classifier is a clinical problem. We now describe how we define a problem. Medical concepts are first extracted using the MedLEE natural language processor and tagged using Concept Unique Identifiers (CUIs) defined in the National Library of Medicine’s Unified Medical Language System (UMLS). MedLEE labels concepts into types, including “problem,” based on several criteria, among them their UMLS types. The MedLEE problem labeling, however, can be overly broad (e.g., the top-level concept C0013428 Disease is tagged a problem). Thus, we also relied on the SNOMED-CT Problem List Subset (PLS) developed by the Veterans Health Administration and Kaiser Permanente. The PLS contains a list of manually selected problems, and has been shown to be adequate for representing medical conditions15. To avoid missing specific problems, concepts which do not occur in the PLS but have a SNOMED parent in the PLS, were included.
Once the concepts are labeled and problems are identified, we aggregate problems across the dataset.
The SNOMED terminology can represent terms to a finer level of granularity than is required for a summary. To improve likelihood of identifying trends from similar, overly granular problem descriptions, as well as to avoid having virtually synonymous problems repeated in the summary, CUIs were clustered according to their SNOMED proximity. For example, without any grouping method, problems such as C0340305 Inferior Wall Myocardial Infarction, and C0340312 Lateral myocardial infarction NOS are considered by the classifier independently from C0027051 Myocardial Infarction. With grouping, references to the first two are rolled into the standard MI for purposes of content selection. For each problem in the Renal Corpus, the Expert Summary Note Corpus, or the Expert Summaries, the SNOMED parent problems are identified also existing in the corpora and the PLS. The process was repeated until reaching a concept that was not in the PLS. Optionally, upward mapping was terminated if the parent problem’s frequency of use was less than that of the child. Upon arrival at a terminal ancestor, the original problem is mapped to the ancestor for all analyses. For a summary, the highest problem occurring in the patient corpus would be presented.
Features:
We experimented with a wide range of features. Several were based on traditional information retrieval statistics, such as inverse document frequency (IDF) and patient term frequency (TF). Others we derived based on hypotheses of usage patterns in patient data. For example, short duration problems like a mild bacterial infection or low-grade trauma will be not have lasting relevance, whereas a chronic problem such as diabetes mellitus or a highly relevant acute problem such as a myocardial infarction are more likely to be carried forward from old notes into a summary. Therefore assessing note usage features such as persistence should be generally indicative of problem relevance, whether acute or chronic in nature.
Content selection features are based on problem usage observed within the corpus, either at the patient level or patient-independent features. A few features are assessed both at the patient level and corpus-wide. For example, the duration of a CUI for a patient may provide insight to how severe the problem has been for the patient. Problem duration is also averaged across all patients and included in the classifier features as an indicator of how complex the problem tends to be, to provide insight to the classifier that it may be relevant, even if it has not been present for long in the current patient. Some additional features are based on external datasets.
Patient-Independent Features
UMLS CUI: the simplest feature assessed on any problem, the CUI string itself
- IDF: average inverse document frequency of the problem in notes containing it
Corpus duration: days from the first reference to the last, averaged across all patients
Corpus duration percent: duration over days to last patient note, averaged across all patients
Corpus persistence: percent of following notes referring to the problem, across all patients
Corpus TF: percent of problem references in a note to this particular CUI, across all patients
Corpus PMH frequency: percent of PMHs mentioning the problem, across all patients
Semantic types: all UMLS types to which the CUI is classified
Patient-Specific Features
- Patient frequency: term frequency calculated on positive problem references across patient’s notes
Duration: days from the first instance to the last
Duration percent: duration divided by days from the first instance to the date of the last patient note
Persistence: the percentage of notes from the first referencing the CUI to the last patient note which mention the CUI
- Term frequency: much like Patient Frequency, but note centric as standard TF would be calculated
- Average TF-IDF: average TF-IDF across all notes
Average TF-IDF density: average TF-IDF of the CUI across only notes containing it
Days since first mention: CUI age - simply the number of days elapsed from the first note the CUI was mentioned until the date of the last note
Percent negated: the percent of CUI instances MedLEE found to be negated
Last negated: if the last instance was negated
First section: the first section where seen
Most common section: the most common section in which the CUI was seen
Sections: sections in which the problem was seen
Note types: note types where the problem is seen
Model Evaluation:
Patient problems and features were used to build classifiers using two Weka16 algorithms: Naïve Bayes and J48, Weka’s implementation of the C4.517 decision tree. These were selected in advance as both are generally accepted as meaningful and accurate classifiers with different strengths. Both classifiers were trained on problem instances observed in the Renal Note Corpus. Expert Summaries then served as an independent test set for evaluation. For each of the four evaluation patients, the patient’s notes were filtered from the Renal Corpus, the classifier was trained in all remaining patients, and then for each problem of the evaluated patient the classifier was queried for problem inclusions and the result was compared with problem inclusion.
Standard information retrieval metrics were used for assessing the classifier built: precision, recall, f-measure. The 95% Confidence Interval (CI) were calculated to ensure statistically significant differences. From this, we report an adjusted TCR, which is forced to 0 if the error rate of the results shows no significant difference to the baseline.
Input Section Analysis:
This experiment was performed to assess whether the summary could be improved by limiting assessed problems to instances from particular source sections. For each of the 12 primary sections used, classifiers were built on problems referenced at least once in the section in question of the current patient’s notes. Results were based on using a Naïve Bayes classifier and problems were filtered with the PLS and grouped using the frequency-limited PLS grouper.
To show whether a concept’s source section has a major effect on the relevance of the summary, corpus data and model representations are built with concepts extracted only from specific source sections or combinations of sections.
Feature Selection:
Chi-Square (χ2) ranking was used to evaluate the relative importance of each feature, and classifiers were built using the top five and top ten features. Best-first and Genetic feature selection algorithms were also assessed for identification of optimal feature sets.
Negated Finding Filtering:
To evaluate removing negated problems, problems where over 50% of instances were negated or where the final instance was negated (as proposed by Meystre18) were filtered and classifiers were rerun.
Results
Input Section Analysis:
Limiting the sections from which classifier training data were drawn was performed on 11 individual sections, the combination of the top five performing sections. In all cases, analysis yielded significant decreases to F-Measure.
Feature Selection:
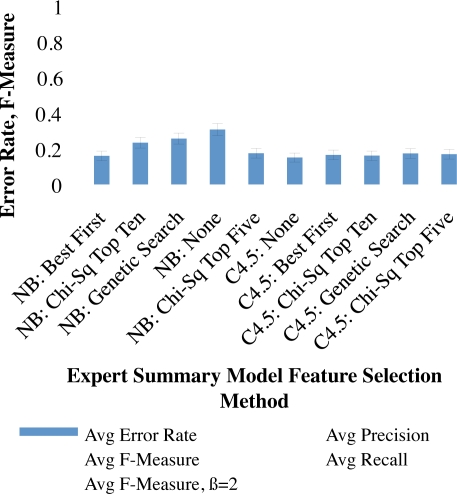
χ2 ranking ordered top features as follows: Patient Frequency, TF, IDF, UMLS CUI, Corpus Duration Percent, Corpus Persistence, Corpus PMH Frequency, Duration Pct, Duration, and Corpus TF. Models with the top five and top ten features were incorporated into analyses along with best-first and genetic search feature selection algorithms. Error-rate, precision, recall, F-measure and F2-measure for each of these are compared against using no feature selection algorithm in Figure 1.
Figure 1:
Evaluation with different feature-selection methods
Filtering Negated Findings:
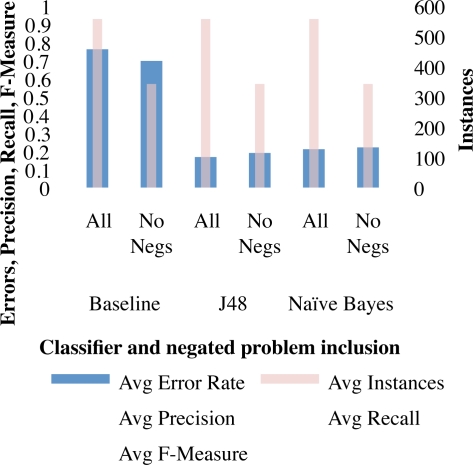
As shown in Figure 2, the application of a simplistic filter to remove negated problems resulted in increased precision and recall using both classifiers as well as the baseline.
Figure 2:
Findings when applying a basic filter for omitting generally negated problems
Overall Evaluation:
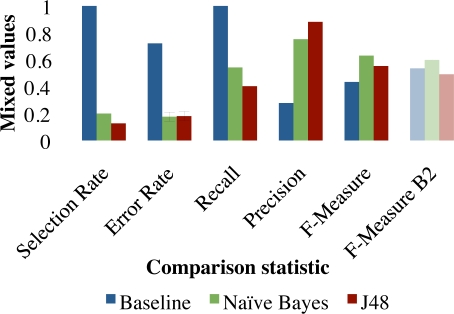
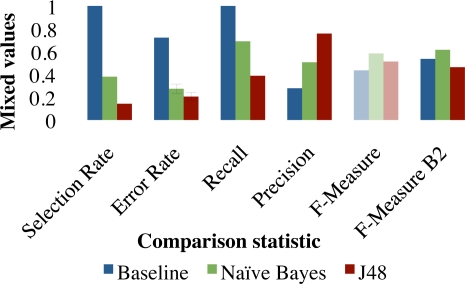
Preceding analyses showed input section restrictions to be unhelpful, while our method for negated problem filtering was. Which feature selection method is best is dependant on the cost of omission. Figures 3 and 4 compare selection rates, error rates, precision, recall, F-measure and F2-Measure for the baseline of selecting 100% of problems as well as filtering problems with the Naïve Bayes and C4.5 classifiers. If we assume equal importance of precision and recall, using Best-First feature selection helped to omit extraneous problems from the summary, the results of which are shown in Figure 3. When recall is assumed to be twice as important as precision and we optimize on the F2-Measure, results were better when not using any feature selection. Results without feature selection are shown in Figure 4.
Figure 3:
Evaluation with the best parameter using Best-First feature selection
Figure 4:
Evaluation with the best parameter set discovered above
Discussion
The input section analysis suggested that problem extraction may not be limited to particular sections. Our method for filtering generic problems and grouping granular problems had little effect, though this warrants additional research. Feature evaluation with the χ2 ranking showed the importance of a few features on the success of this model. The model included some attributes for assessing note-based usage patterns as would be done in traditional information retrieval, while others assessed time-based usage patterns such as Duration. Note-based usage patterns performed significantly better, though this may well be a consequence of a relatively short time span of the Renal Corpus. It was found that patient-specific and patient-independent features were both relevant.
Issues of coding granularity created problems such that two concepts seen in text with very similar meaning were meaning were given different codes and therefore treated independently in the model. This served to distract the classifier and evaluation by confounding reference matching between notes and expert summaries. A detailed experiment was performed to group related concepts, but results were inconclusive. This problem warrants significant additional research.
Conclusions
We presented an approach to selecting problems relevant for a clinical summary in patient notes. Evaluation shows relevance classification accuracy as high as 82% and an F-measure of 0.62. These results are promising and suggest that patient summaries can be reliably generated for clinical purposes.
Acknowledgments
This work was supported by National Library of Medicine grants N01-LM07079 (TTVV) and R01 LM010027 (NE). Thanks to Dr. Carol Friedman for the use of MedLEE. MedLEE development is funded by NLM R01 LM007659 and R01 LM008635.
References
- 1.Van Vleck TT, Wilcox A, Stetson PD, Johnson SB, Elhadad N. Content and structure of clinical problem lists: a corpus analysis. Proc AMIA Annu Fall Symp. 2008:753–7. [PMC free article] [PubMed] [Google Scholar]
- 2.Weed LL. Medical records that guide and teach. N Engl J Med. 1968;278(12):652–7. doi: 10.1056/NEJM196803212781204. concl. [DOI] [PubMed] [Google Scholar]
- 3.Meystre S, Haug P. Randomized controlled trial of an automated problem list with improved sensitivity. IJMI. 2008;77(9):602–12. doi: 10.1016/j.ijmedinf.2007.12.001. [DOI] [PubMed] [Google Scholar]
- 4.Solti I, Aaronson B, Fletcher G, Solti M. Building an Automated Problem List Based on Natural Language Processing: Lessons Learned in the Early Phase of Development. Proc AMIA Annu Fall Symp. 2008 [PMC free article] [PubMed] [Google Scholar]
- 5.Rassinoux A, Miller R, Baud R, Scherrer J. Modeling just the important and relevant concepts in medicine for medical language understanding: a survey of the issues. Proc IMIA Working Group. 6:19–22. [Google Scholar]
- 6.Humphreys B, Lindberg D, Schoolman H, Barnett G. The Unified Medical Language System: an informatics research collaboration. JAMIA. 1998 doi: 10.1136/jamia.1998.0050001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cote R, Robboy S. Progress in medical information management: Systematized Nomenclature of Medicine (SNOMED) JAMA. 1980 doi: 10.1001/jama.1980.03300340032015. [DOI] [PubMed] [Google Scholar]
- 8.Hahn U, Romacker M, Schulz S. How knowledge drives understanding—matching medical ontologies with the needs of medical language processing. Artificial Intelligence In Medicine. 1999 doi: 10.1016/s0933-3657(98)00044-x. [DOI] [PubMed] [Google Scholar]
- 9.Zweigenbaum P, Bouaud J, Bachimont B, Charlet J. Evaluating a normalized conceptual representation produced from natural language patient discharge summaries. Proc AMIA Annu Fall Symp. 1997 [PMC free article] [PubMed] [Google Scholar]
- 10.Aronson A. Effective mapping of biomedical text to the UMLS Metathesaurus: the MetaMap program. Proc AMIA Annu Fall Symp. 2001 [PMC free article] [PubMed] [Google Scholar]
- 11.Chapman W, Bridewell W, Hanbury P, Cooper G, Buchanan B. A Simple Algorithm for Identifying Negated Findings and Diseases in Discharge Summaries. Journal of biomedical informatics. 2001;34:301–10. doi: 10.1006/jbin.2001.1029. [DOI] [PubMed] [Google Scholar]
- 12.Friedman C, Hripcsak G, Shagina L, Liu H. Representing Information in Patient Reports Using Natural Language Processing and the Extensible Markup Language. J Am Med Inform Assoc. 1999 doi: 10.1136/jamia.1999.0060076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Elhadad N, McKeown K. Towards generating patient specific summaries of medical articles. Proc of NAACL Workshop on Automatic Summarization. 2001 [Google Scholar]
- 14.Mendonça EA. Using Automated Extraction from the Medical Record to Access Biomedical Literature. Columbia Dissertation. 2002;134 [Google Scholar]
- 15.Mantena S, Schadow G. Evaluation of the VA/KP Problem List Subset of SNOMED as a Clinical Terminology for Electronic Prescription Clinical Decision Support. Proc AMIA Annu Fall Symp. 2007;2007:498. [PMC free article] [PubMed] [Google Scholar]
- 16.Witten IH, Frank E, Trigg L, Hall M, Holmes G, Cunningham SJ. Weka: Practical Machine Learning Tools and Techniques with Java Implementations. 2002.
- 17.Quinlan J. C45: Programs for machine learning. 1993.
- 18.Meystre S, Haug P. Automation of a problem list using natural language processing. BMC Med Inform Decis Mak. 2005;5(30) doi: 10.1186/1472-6947-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]