Abstract
As the use of detailed clinical data expands for strategic planning, clinical quality measures, and research, the quality of the data contained in source systems, such as electronic medical records, becomes more critical. Methods to quantify and monitor clinical data quality in large operational databases involve a set of predefined data quality queries that attempt to detect data anomalies such as missing or unrealistic values based on meta-knowledge about a data domain. However, descriptive data elements, such as patient race, cannot be assessed using these methods. We present a novel approach leveraging existing intra-institutional databases with differing data quality for the same data element to quantify data quality for descriptive data. Using the concept of a relative gold standard, we show how this method can be used to assess data quality in enterprise clinical databases.
Introduction
Health care organizations are moving rapidly to implement comprehensive enterprise data warehouses (EDWs) that provide an institutionally defined “source of truth” for operational, quality and patient safety measures; strategic decision-making, and clinical research.1–6 High quality data are critical to defining robust enterprise definitions and measures of patient conditions and clinical outcomes.
Dedicated electronic data capture systems developed for clinical trials have sophisticated point-of-entry data validation processes to detect potential data quality issues during data entry.7 Examples of good data management features used in dedicated clinical trials systems include pick-lists rather than free-text, range checks on numeric and calendar values, and consistency checks based on one or more data elements contained in a form or across multiple values in the database.8
Enterprise health care data repositories obtain data from multiple operational clinical and financial systems. In many cases, the source system for electronic clinical data in an EDW comes from an electronic medical record (EMR) system. Modern EMRs contain a growing array of tools to capture data directly from busy clinical care providers.9–11 Although features contained in research-oriented data capture tools are found in EMRs, most documentation tools have focused on improving the efficiency of data entry rather than on ensuring data quality.12 Thus, data contained in most operational systems have been shown to have significant completeness, accuracy, and quality issues.13–16
Assessing the quality of data within traditional business-oriented systems has a substantial body of literature and best-practices.17–19 Assessing the quality of data within an EMR is an unsolved informatics challenge. Current assessment methods usually involve creating data quality queries that examine fields for missing, abnormal or unbelievable values, or inconsistent relationships between pairs (“double-checks”) or triplets (“triple-checks”) of variables. Each EMR or EDW team creates a set of quality check queries based on the data elements that are important to the enterprise, usually for reporting or analysis. When data anomalies are detected, manual chart reviews are usually performed to validate the data quality issue. These findings are provided back to the parties responsible for collecting the original data for corrective action.
A key difficulty in assessing data quality is the lack of a gold standard, especially for qualitative clinical or demographic features. In this effort, we have focused on patient race, which, in our operational EMR, often is listed as “unknown.” Because an individual may be a member of any value for race, this data element is impossible to validate using the “abnormal value detection” quality assessment method. Our approach leverages differences in data quality across institutional data resources, using a data source deemed to be more accurate as a relative gold standard. The concept of a relative gold standard is the key insight which allows us to use one internal data source to assess the data quality in another internal data source. In the following sections, we present our conceptual model followed by methods, findings, and discussion, including limitations.
Conceptual Model: Relative Data Quality
We assume that various databases within an enterprise have different levels of data quality, driven by the amount of effort invested by the data owners to maintain data quality and integrity. In many institutions, numerous specialized data sources, with narrow scope supported by dedicated data owners, capture mission-critical or research-oriented data elements. Derisively called “boutique databases” and often scorned by centralized IT due to concerns about data management practices and data security, these systems are closely maintained by a small group of highly motivated individuals who view this data source as central to the success of their operational, regulatory, or research program. These specialized databases may contain highly accurate data on well-defined patient sub-populations that are pertinent to the local group. In this setting and only for patients within the specialized database, data elements recorded in these systems can be of extremely high quality because of the critical nature of these data values to the data owners.
An institutional data source that is known or thought to have higher data quality in a data domain is considered a relative gold standard for data quality when compared to other sources that contain the same data domain. We use the term relative gold standard for data quality to acknowledge the reality that even with an assumed higher level of data quality, we expect that the relative gold standard also contains errors. We assume that the error rate in a relative gold standard is substantially lower than in systems that do not invest in the same effort to ensure data quality in that data domain. The relative gold standard applies to a specific data domain in a data source. The same data source may not be a relative gold standard in other data domains that it contains. An example of a relative gold standard that we have used to examine this conceptual model is the patient demographics information contained within the Hematology, Oncology, Bone Marrow Transplant database (HOB-DB) at The Children’s Hospital, Denver.
The HOB-DB contains detailed demographic, clinical and outcomes data on 4,191 patients, of which 1,874 patients were consented to one or more clinical protocols. For consented patients, the data contained within the HOB-DB is collected by dedicated research coordinators and data collection personnel with oversight by a full time database coordinator. The data are heavily exploited to support HOB clinical operations and an extensive array of clinical research projects. In addition, the database is used to report vital statistics to local, state and national databases and to funding agencies. Because of the critical role of the HOB-DB on multiple departmental missions, substantial efforts are expended to ensure an extremely high level of data quality, including detailed data collection procedures and extensive validation checks that are not possible to perform within an operational clinical system such as an EMR.
One data element that is deemed especially critical to the HOB investigators is the accurate assessment and recording of patient race for all consented patients, using the NIH race categories. This is a key element for NIH grants and for clinical trials recruitment. Substantial efforts have been put in place to ensure that HOB personnel are trained to assess and record this data element. Thus, race data are highly accurate in the HOB-DB, making the HOB-DB a relative gold standard for patient race.
We consider the HOB-DB to be a relative gold standard compared to the EMR for patient race. With this assumption, we compared the correspondence in the values for patient race assigned to the subset of patients found in both databases. We assume that patients common to the HOB-DB and the EMR have roughly the same data quality for race in the EMR as do patients not in the HOB-DB so that the agreement or disagreement rate in HOB-DB patient races is a reasonable estimate of the agreement or disagreement rate across the EMR. This assumption will be examined further in the Discussion.
We do not assume that all data domains in the HOB-DB are superior in data quality compared to the EMR. A given data source may be a relative gold standard in only a few domains based on specific business drivers that motivate data owners to expend special attention on the accuracy of these domains.
Methods
In this example, we seek to quantify data quality for race in the EMR by comparing the concordance and discordance of recorded values for patient race between two data sources, the EpicCare® EMR and the HOB-DB at The Children’s Hospital, Denver. Data on patient race was extracted for all patients with one or more billed encounters in EPIC and all consented patients in HOB-DB.
Race value domains used in the two data sources were mapped to a common set of race terms. The mapping from each source to the common terms was determined manually by the research team (BBE). Data accuracy measures are described using the HOB-DB as the relative gold standard compared to the EMR data set. For patients who appeared in both data sets, the inter-rater agreement was calculated using Cohen’s kappa coefficient.20 All analyses were done using SPSS Version 17; IBM Corp.
This study was approved by the Colorado Combined Multiple Institutional Review Board (COMIRB) as an exempt study.
Results
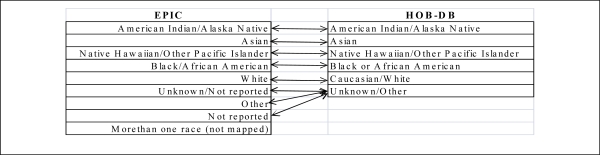
There were 478,403 unique patients with one or more billed encounters in the TCH Epic EMR. There were 1,864 unique consented patients in the HOB-DB. There were 1,192 unique patient races that were common and mapped between both data sources. The EMR contained 9 distinct race codes and the HOB-DB contained 6 distinct race codes. Table 1 highlights the mapping between the race domain values across the two data sources.
Table 1:
Race domain values cross-mappings between EPIC EMR and Hematology/Oncology/Bone Marrow Transplant research database.
Table 2 provides the breakdown of matches and mismatches across the HOB-DB and EMR. Because HOB-DB was considered the relative gold standard, matches in Table 2 are based on the race assignment found in the HOB-DB. Cells in gray along the 45-degree diagonal denote exact matches for each of the six HOB-DB race codes. Off-diagonal cells are race mismatches. Overall, only 68% (810/1192) entries matched on mapped race codes. Cohen’s kappa for the overall agreement rate was 0.26 (95% CI: 0.22–0.30) indicating a very low agreement rate compared to chance agreement.
Table 2:
Matches and Mismatches by Race: HOB-DB (gold standard) versus Epic EMR.
| HOB-DB (“Relative Gold Standard”) | ||||||||
|---|---|---|---|---|---|---|---|---|
| Whte | Black | Asian | AIAN | NHPI | Unkn/Other | |||
| EPIC | White | 720 | 3 | 3 | 3 | 1 | 4 | 734 |
| Black | 4 | 22 | 0 | 0 | 0 | 0 | 26 | |
| Asian | 1 | 0 | 16 | 0 | 0 | 0 | 17 | |
| AIAN | 4 | 0 | 0 | 9 | 0 | 0 | 13 | |
| NHPI | 0 | 0 | 0 | 0 | 2 | 0 | 2 | |
| Unkn/Other | 342 | 5 | 8 | 4 | 0 | 41 | 400 | |
| 1071 | 30 | 27 | 16 | 3 | 45 | 1192 | ||
AIAN = American Indian/Alaskan Native; NHPI = Native Hawaiian or Other Pacific Islander. Gray cells denote agreement between HOB-DB and EPIC reported race values.
Of the 1,192 race entries in the common population, the HOB-DB contained 1,071 (90%) entries with a race definition of “White/Caucasian”. Of these White/Caucasian races in HOB-DB, only 720 (67%) were also identified as White/Caucasian in the EMR.
Similar matching frequencies were seen for Black (22/30=73%), Asian (16/27=59%) and American Indian or Alaskan Native (9/16=56.
Overall, the HOB-DB identified 45 entries (3% of all races) as “Unknown/Other” whereas the EMR marked 400 entries as “Unknown”, “Not Reported” or “Other” (33% of races), a roughly 10-fold difference in the number of unknowns between the two databases. Thus the HOB-DB provided more specific race assignments than the EMR in 359 races. Of these 359 entries, HOB-DB listed 342 entries as “White.” Of the 45 races listed as unknown/other in HOB-DB, 41 were also listed as unknown/other in the EMR.
Discussion
Using the HOB-DB as a relative gold standard, we found that only 68% of entries for race common to both the HOB-DB and the EMR matched on race code. A large proportion of the mismatches were due to a more specific race code assigned in the HOB-DB when the EMR assigned “Unknown”, “Not Reported”, or “Other.” This finding of increased specificity adds support for the assessment that HOB-DB is a relative gold standard compared to the current EMR for this variable.
Our approach provides a method for assessing data quality in value domains that are not amenable to traditional range or cross-validation checks. There are no surrogate markers for race that can be used to determine the reasonableness of a value for a patient. Traditional data quality queries can only examine the number of null values or the number of “Unknown/Not Reported” values. Using a relative gold standard, the quality of recorded values for patient race can be estimated, including an expected rate for the number Unknown/Not Reported values. Identifying a relative gold standard from a specialized data source and using those data as a data quality probe can be expanded to additional data domains such as death dates, ethnicity, and problem lists.
Using a second source of data to compare or validate a data source is not new, especially in the research context. For example, Raebel et. al compared gestational age and admission dates across two separate data sets, the Kaiser Permanente Birth Registry and the Prescribing Safely during Pregnancy research dataset .21 Our approach extends this commonly used strategy by highlighting the value of existing, internal (and usually ignored) “boutique” databases that exist within an organization.
Our approach contains a number of assumptions and limitations that require additional exploration before promoting wider adoption. There is no absolute measure of data quality that can be calculated for all data domains. The determination of the data quality in a relative gold standard is a subjective assessment based on meta-knowledge about the proposed relative gold standard data source, such as business drivers and organizational features such as the level of training and oversight applied during data collection. A gold standard could be created by collecting a small amount of data using rigorous data collection protocols to provide a measure of the quality of the relative gold standard.
We assume that the distribution of data errors in the sub-population contained in the relative gold standard is the same as the distribution of data errors in patients not contained in the joint population. In this example, we are assuming that error rate in assigning race in the EMR for 1,192 matching HOB-DB race entries are identical to error rate in assigning race for the remaining 477,211 non-HOB patients, allowing the match/mismatch rate in HOB patients to be an estimation of the match/mismatch rate for race across the entire EMR. If there is a bias to be selectively more (or less) accurate in assigning race to HOB patients by EPIC registration personnel, then this assumption will be incorrect. For example, if hospital personnel are more vigilant in assigning race to HOB patients than to non-HOB patients in the EMR, then the degree of mismatches seen in HOB patients would under-estimate the actual degree of mismatches in the EMR.
We do not know the true error rate in the relative gold standard, although we described a procedure for obtaining an estimate. If the data quality in the relative gold standard is relatively poor, then the kappa coefficient against the HOB patients will be difficult to interpret.
Our methodology requires an accurate cross-map between the value sets across the two databases. The poorer the correspondence, the higher will be the proportion of mismatches due to an incorrect or incomplete mapping between the value sets. Issues with cross-mapping could result in either under-estimating or over-estimating the true quality of the data source being compared to the relative gold standard.
The approach can be extended to assess the impact of data quality improvement efforts by breaking the data to be compared into a “before” and “after” data set and calculating agreement rates in both time intervals. If data quality improvement efforts were successful, one would expect to see an increase in the kappa statistic after the intervention. However, if the improvement project results in the “after” data having greater accuracy than the relative gold standard, a decrease in the kappa statistic could be incorrectly interpreted as poor data quality in the EMR rather than in the relative gold standard, would no longer a viable relative gold standard.
The objective of measuring data quality is to provide a means for detecting when data quality issues exist so that data quality improvement efforts can be targeted. Any automated method, even if not perfect (e.g. using “relative” gold standards), is more likely to be adopted than methods that depend on random manual chart audits. We have shown that a useful assessment of quality can be made with reference only to a relative gold standard.
We have emphasized that a data source that contains high quality data usually has internal or external drivers which cause the data owners to invest more resources in ensuring the quality of their data, such as grant reporting or research agendas. The recently released final rules for meaningful use could become a significant external driver for improving the quality of data for those elements used to calculate quality indicators or other measures that will be incorporated into the final Meaningful Use assessment rules.22 Meaningful Use requirements are also likely to encourage EMR vendors to focus more efforts on supporting high quality data capture methods in their application’s user interfaces. Our experience has been that data elements used in critical reports undergo improvement in their data quality due to the additional scrutiny applied to these data element by report consumers. Meaningful Use would represent a new opportunity to improve the overall data quality in operational EMRs and EDWs.
Acknowledgments
Dr. Kahn is supported in part by Colorado CTSA grant 1 UL1 RR 025780 from NCRR/NIH and the TCH Research Institute. We thank Zettie Chin-Fong and Pamela Wolfe for comments on an earlier draft.
References
- 1.Kamal J, Silvey SA, Buskirk J, Dhaval R, Erdal S, Ding J, et al. Innovative applications of an enterprise-wide information warehouse. AMIA Annu Symp Proc. 2008;1134 [PubMed] [Google Scholar]
- 2.Chute CG, Beck SA, Fisk TB, Mohr DN. The Enterprise Data Trust at Mayo Clinic: a semantically integrated warehouse of biomedical data. J Am Med Inform Assoc. 2010;17(2):131–135. doi: 10.1136/jamia.2009.002691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Murphy SN, Weber G, Mendis M, Gainer V, Chueh HC, Churchill S, et al. Serving the enterprise and beyond with informatics for integrating biology and the bedside (i2b2) J Am Med Inform Assoc. 2010;17(2):124–130. doi: 10.1136/jamia.2009.000893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Grant A, Moshyk A, Diab H, Caron P, de Lorenzi F, Bisson G, et al. Integrating feedback from a clinical data warehouse into practice organisation. Int J Med Inform. 2006;75(3–4):232–239. doi: 10.1016/j.ijmedinf.2005.07.037. [DOI] [PubMed] [Google Scholar]
- 5.Resetar E, Noirot LA, Reichley RM, Storey P, Skiles AM, Traynor P, et al. Using business intelligence to monitor clinical quality metrics. AMIA Annu Symp Proc. 2007;1092 [PubMed] [Google Scholar]
- 6.Ferranti JM, Langman MK, Tanaka D, McCall J, Ahmad A. Bridging the gap: leveraging business intelligence tools in support of patient safety and financial effectiveness. J Am Med Inform Assoc. 2010;17(2):136–143. doi: 10.1136/jamia.2009.002220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Spilker B. Guide to Clinical Trials. New York, NY: Raven Press; 1991. [Google Scholar]
- 8.Society for Clinical Data Management Good Clinical Data Management Practices. 2009 [Google Scholar]
- 9.Johnson KB, Ravich WJ, Cowan JA. Brainstorming about next-generation computerbased documentation: an AMIA clinical working group survey. Int J Med Inform. 2004;73(9–10):665–674. doi: 10.1016/j.ijmedinf.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Schnipper JL, Linder JA, Palchuk MB, Einbinder JS, Li Q, Postilnik A, et al. “Smart Forms” in an Electronic Medical Record: documentation-based clinical decision support to improve disease management. J Am Med Inform Assoc. 2008;15(4):513–523. doi: 10.1197/jamia.M2501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Whipple NN, Palchuk MB, Olsha-Yehiav M, Li Q, Middleton B. Supporting CMT and user customization in Clinical Documentation templates. AMIA Annu Symp Proc. 2007;1153 [PubMed] [Google Scholar]
- 12.Rosenbloom ST, Crow AN, Blackford JU, Johnson KB. Cognitive factors influencing perceptions of clinical documentation tools. J Biomed Inform. 2007;40(2):106–113. doi: 10.1016/j.jbi.2006.06.006. [DOI] [PubMed] [Google Scholar]
- 13.Aronsky D, Haug PJ. Assessing the quality of clinical data in a computer-based record for calculating the pneumonia severity index. J Am Med Inform Assoc. 2000;7(1):55–65. doi: 10.1136/jamia.2000.0070055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stein HD, Nadkarni P, Erdos J, Miller PL. Exploring the degree of concordance of coded and textual data in answering clinical queries from a clinical data repository. J Am Med Inform Assoc. 2000;7(1):42–54. doi: 10.1136/jamia.2000.0070042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hogan WR, Wagner MM. Accuracy of data in computer-based patient records. J Am Med Inform Assoc. 1997;4(5):342–355. doi: 10.1136/jamia.1997.0040342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wagner MM, Hogan WR. The accuracy of medication data in an outpatient electronic medical record. J Am Med Inform Assoc. 1996;3(3):234–244. doi: 10.1136/jamia.1996.96310637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Maydanchik A. Data Quality Assessment. Bradley Beach, NJ: Technics Publications; 2007. [Google Scholar]
- 18.Redman TC. Data Quality: The Field Guide. Boston: Digital Press; 2001. [Google Scholar]
- 19.Batini C. Data Quality: Concepts, Methodologies and Techniques. Berlin: Springer; 2006. [Google Scholar]
- 20.Cohen J. A coefficient of agreement for nominal scales. Educational and Psychological Measurment. 1960;20(1):37–46. [Google Scholar]
- 21.Raebel MA, Ellis JL, Andrade SE. Evaluation of gestational age and admission date assumptions used to determine prenatal drug exposure from administrative data. Pharmacoepidemiol Drug Saf. 2005;14(12):829–836. doi: 10.1002/pds.1100. [DOI] [PubMed] [Google Scholar]
- 22.Centers for Medicare & Medicaid Services Electronic Health Record Incentive Program [Internet] 2010. [cited 2010 Jul 14];Available from: http://www.ofr.gov/OFRUpload/OFRData/2010-17207_PI.pdf.