Abstract
This paper presents a novel approach to learning semantic classes of clinical research eligibility criteria. It uses the UMLS Semantic Types to represent semantic features and the Hierarchical Clustering method to group similar eligibility criteria. By establishing a gold standard using two independent raters, we evaluated the coverage and accuracy of the induced semantic classes. On 2,718 random eligibility criteria sentences, the inter-rater classification agreement was 85.73%. In a 10-fold validation test, the average Precision, Recall and F-score of the classification results of a decision-tree classifier were 87.8%, 88.0%, and 87.7% respectively. Our induced classes well aligned with 16 out of 17 eligibility criteria classes defined by the BRIDGE model. We discuss the potential of this method and our future work.
INTRODUCTION
Clinical research eligibility criteria are an important section of clinical research protocols that specify the mandatory characteristics of clinical research participants. Often, a criterion is just a short phrase or sentence fragment, such as “Age: 18–80” or “No DSM-IV diagnosis other than schizophrenia”. Computable eligibility criteria have been desired for its promising applications in automatically matching patients to clinical trial opportunities or results. Knowledge representation, a popular method to achieve computable eligibility criteria, often introduces laborious manual efforts in identifying semantic classes of eligibility criteria as well as frequent modeling variations. Our prior study has shown that clinical research eligibility criteria have been classified in varied ways for different applications, which is a big barrier to standardization of clinical research eligibility criteria models.1
Automated induction of semantic classes from text has been frequently studied to improve knowledge acquisition efficiency2–4. Most of the prior works were focused on identifying semantic word classes. In contrast, we aim to semi-automatically identify sentence classes of eligibility criteria with three rationales. First, each eligibility criterion sentence is an independent patient characteristic. Automatic sentence classification can enable us to compute the coverage, distribution, and frequency of patient characteristics in clinical research eligibility criteria. Second, when building a knowledge base of computable eligibility criteria, automatic sentence classification can significantly reduce manual efforts for knowledge acquisition (e.g., categorizing eligibility criteria and selecting encoding templates). Third, we hypothesize that corpus-based knowledge acquisition method is more efficient and systematic than classic approaches that lean heavily on domain expertise for knowledge representation, and can help standardize shared knowledge models to support scalable natural language processing systems. Semi-automatic sentence classification is an initial step toward corpus-based knowledge acquisition of clinical research eligibility criteria.
Clustering methods are popular solutions to semantic class learning for various applications, such as ontology development5, content organization6, and thesaurus construction7. The prior works typically used the “bag of words”8 as learning features. However, this approach does not recognize multiword terms, which are typical in medicine. Our experiments also showed that about 70% of terms in the eligibility criteria corpus were unique.9 A feature space using the “bag of words” could be too large to be efficient. We hypothesize that the use of The Unified Medical Language Systems (UMLS) semantic types can decrease the feature space and achieve satisfactory machine learning efficiency. We have previously developed a UMLS-based semantic lexicon9 and a semantic annotator so that each concept in eligibility criteria can be annotated by one of the 135 UMLS semantic types or one of the 8 new types created for the eligibility criteria text.
Therefore, in this paper, we present a novel approach to inducing semantic classes, which uses the clustering results of semantic features represented by the UMLS semantic types to identify sentence classes with minimal user input for cluster labeling.
METHOD
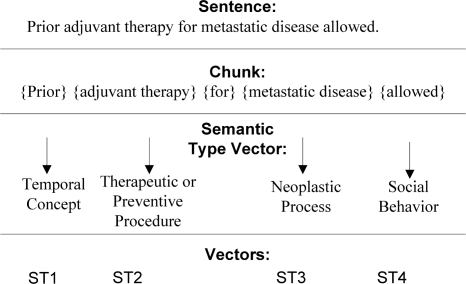
From The Clinicaltrials.gov10, we downloaded about 3,400 randomly selected sample eligibility criteria to carry out this study. Each eligibility criterion was parsed and converted into a UMLS semantic type vector using the steps shown in Figure 1.
Figure 1:
Criteria Semantic Feature Representation.
We used the Hierarchical Clustering Explorer11 to generate clusters of similar eligibility criteria, where the similarity was measured by the Pearson Correlation Coefficient. For sentences X and Y with annotation vectors xi, yi, the similarity C (X ,Y) can be calculated as:
| (2) |
where i ={1, 2, 3 ···k} , K was the number of UMLS semantic types (K=117). We also used the average linkage method to compute the similarity between clusters. If there were clusters O, P and their belonging sentences Xi ,Yj, the similarity (linkage) between clusters L(O, P) is computed as:
| (3) |
where |O|, |P| were the sizes of the two clusters.
Two subsequent manual steps were taken to adjust cluster granularity and to interpret and label clusters. We sorted the hierarchically linked criteria sentences by their semantic similarities and selected a similarity threshold to cut the linked sentences into 41 initial clusters. The similarity threshold was a scale running from 0 to 1. A low similarity threshold would lead to too many groups, while a high similarity threshold would produce too few groups. We set the threshold to be 0.75, which meant that criteria sentences should to be ≥ 75% similar to form a cluster. Since the use of automatic similarity threshold does not always result in clusters with desired granularity for a particular application, we manually reviewed and merged the 41 clusters into 27 distinct semantic classes based on the manually judged semantic similarity between the clusters (e.g., the clusters contains similar eligibility criteria) and similarity in the patterns of their associations with the UMLS semantic types (see Table 1 below).
Table 1.
Associated UMLS semantic types for selected classes.
| Semantic Class | Top 3 Semantic Types | Freq % |
|---|---|---|
| Addictive Behavior | Behavior Problem | 0.28 |
| Temporal Concept | 0.11 | |
| Pharmacologic Substance | 0.07 | |
| Allergy | Pharmacologic Substance | 0.23 |
| Allergy | 0.22 | |
| Qualitative Concept | 0.12 | |
| Consent | Regulation or Law | 0.26 |
| Finding | 0.15 | |
| Functional Concept | 0.11 | |
| Device | Medical Device | 0.34 |
| Therapeutic or Preventive Proce- | 0.08 | |
| Body Part Organ | 0.07 | |
| Diagnostic or Lab Results | NUMERAL | 0.17 |
| SYMBOL | 0.12 | |
| UNIT | 0.11 | |
| Disease, Symptom and Sign | Disease or Syndrome | 0.24 |
| Qualitative Concept | 0.13 | |
| Functional Concept | 0.06 | |
| Enrollment in other studies | Other Study | 0.16 |
| Research Activity | 0.12 | |
| Qualitative Concept | 0.09 | |
| Ethnicity | Ethnicity | 0.37 |
| Qualitative Concept | 0.20 | |
| Population Group | 0.14 |
RESULTS
1. Induced Semantic Classes and their Groups
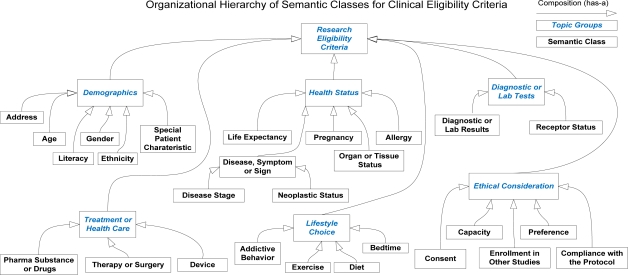
Figure 2 shows the 27 semi-automatically induced semantic classes of clinical research eligibility criteria with minimal user input. They form 6 exclusive topic groups, which are Demographics (e.g., age or gender), Health Status (e.g., disease or organ status), Treatment or Health Care (e.g., drug), Diagnostic or Lab Tests (e.g., creatinine), Ethical Consideration (e.g., willing to consent), and Lifestyle Choice (e.g. diet or exercise). Each of the 27 classes is a member of one of the 6 topic groups.
Figure 2.
| Class | Example Criterion | Class | Example Criterion |
|---|---|---|---|
| Addictive Behavior | Smokes at least 20 cigarettes per day (1 pack per day) | Ethnicity | Patients must be self-identified as African-Americans. |
| Address | Residence in the study area. | Exercise | Currently engaged in vigorous exercise training. |
| Age | Ages Eligible for Study: 18 Years and older. | Gender | Genders Eligible for Study: Female |
| Allergy | Known sensitivity or contra indication to Brimonidine. | Life Expectancy | Life expectancy of at least 6 months |
| Bedtime | Usual bedtime between 21:00 and 01:00. | Literacy | Subjects must be able to read and write in English. |
| Capacity | Able to take oral medications. | Neoplasm Status | No evidence of metastatic disease in the major viscera. |
| Compliance With Protocol | Ability to comply with research procedures. | Organ or Tissue Status | Patient must have adequate organ function. |
| Consent | Signed written informed consent. | Patient Preference | Willingness to come to the health facility for next month |
| Device | Permanent pacemaker or defibrillator. | Pharma Substance or Drug | Patients taking greater than 81mg aspirin daily. |
| Diagnostic or Lab Results | Abnormal laboratory results such as : Hb<8 g/dl | Pregnancy-related Activity | Pregnant or lactating women. |
| Diet | Ingestion of grapefruit or grapefruit juice within 1 week. | Receptor Status | Hormone receptor status not specified |
| Disease Stage | Soft tissue sarcoma chemosensible, stage IV. | Special Patient Characteristic | This protocol is approved for prisoner participation. |
| Disease, Symptom and Sign | Documented coronary heart disease | Therapy or Surgery | Previous radioimmunotherapy within 12 week. |
| Enrollment in Other Studies | Simultaneous participation in other therapeutic trials |
Table 1 shows the top 3 frequent UMLS semantic types for some selected classes. For instance, 23% terms in an “Allergy” criterion have the semantic type “Phyarmacologic Substance”, 22% have the semantic type “Allergy”, and 12% have the semantic type “qualitative concept”. We can extend such patterns of frequently associated UMLS semantic types into class-specific regular expressions or parsing rules to help convert clinical eligibility criteria into structured, computable formats. For example, in an informal test of 84 random selected “Diagnostic or Lab Results” criteria, we discovered that 56 (67%) of the criteria can be parsed using a simple “regular expression pattern” specified by the following semantic types: “Clinical Attribute +Symbol +Numeral +Unit”.
2. Classification Accuracy
To evaluate the usefulness of the induced classes, we trained a C4.5 decision tree classifier. Two human raters blind to machine classification results manually labeled the 2,718 eligibility criteria sentences using the 27 classes. They achieved an 85.73% inter-rater agreement and an average of 83.25% agreement with the machine classification result (Figure 3).
Figure 3:
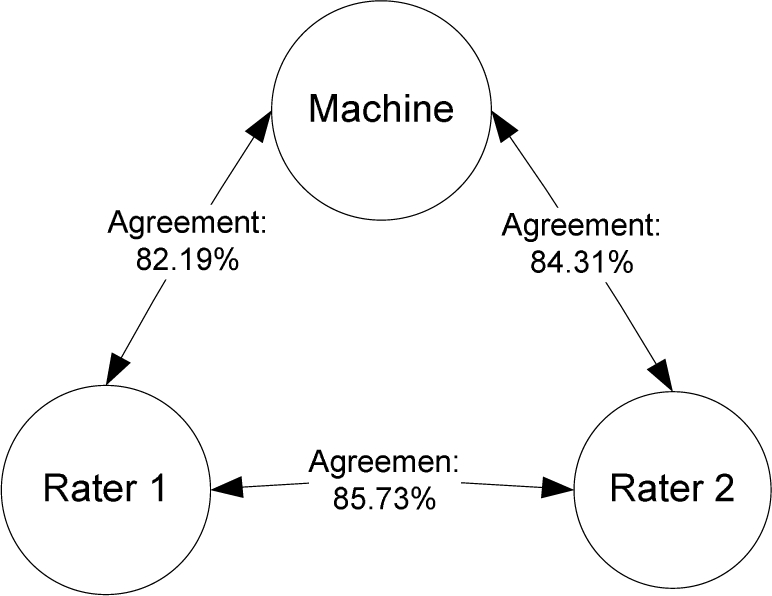
Agreement between two raters and machine
The consensus of the two raters were obtained and used to further train the classifier, whose precision, recall, and F-measure in a 10-fold cross validation test were shown in Table 2. The average F-score for all the classes was 87.7%.
Table 2:
Precision (PR), Recall (RE), and F-Measure (FM of the classification results using manual review by two independent rater as the gold standard (Sorted by F-Score).
| Class Name | Precision | Recall | F-Score |
|---|---|---|---|
| Life Expectancy | 1 | 1 | 1 |
| Allergy | 0.973 | 1 | 0.986 |
| Bedtime | 0.941 | 1 | 0.97 |
| Age | 0.938 | 0.97 | 0.954 |
| Enrollment in other studies | 0.962 | 0.938 | 0.95 |
| Ethnicity | 1 | 0.9 | 0.947 |
| Disease Stage | 0.934 | 0.947 | 0.94 |
| Addictive Behavior | 0.907 | 0.958 | 0.932 |
| Gender | 0.929 | 0.929 | 0.929 |
| Literacy | 0.846 | 1 | 0.917 |
| Disease, Symptom and Sign | 0.894 | 0.912 | 0.903 |
| Exercise | 0.875 | 0.933 | 0.903 |
| Pregnancy , Nursing or Sexual Behavior | 0.893 | 0.903 | 0.898 |
| Diagnostic or Lab Results | 0.906 | 0.873 | 0.89 |
| Pharmaceutical Substance or Drug | 0.864 | 0.904 | 0.884 |
| Address | 0.846 | 0.917 | 0.88 |
| Consent | 0.838 | 0.883 | 0.86 |
| Neoplasm Status | 0.863 | 0.856 | 0.859 |
| Diet | 1 | 0.692 | 0.818 |
| Receptor Status | 1 | 0.671 | 0.803 |
| Therapy or Surgery | 0.783 | 0.764 | 0.773 |
| Patient Preference | 0.778 | 0.745 | 0.761 |
| Capacity | 0.771 | 0.725 | 0.747 |
| Compliance with protocol | 0.8 | 0.671 | 0.73 |
| Organ or Tissue Status | 0.733 | 0.64 | 0.683 |
| Device | 0.65 | 0.672 | 0.656 |
| Special Patient Characteristic | 0.733 | 0.55 | 0.629 |
| Weighted Average | 0.878 | 0.88 | 0.877 |
3. Comparison with the BRIDGE model
There have been manual efforts for developing classes of clinical eligibility criteria. A representative is the BRIDG model 12, which defined 17 eligibility criterion attributes using the consensus of a group of domain experts. We were able to align 16 out of 17 BRIDG attributes with our semantic classes, but also identified 8 classes that were not specified by the BRIDGE model (See Table 3).
Table 3.
Alignment with BRIDGE Criteria Classes
| BRIDG Model | Our 27 Semantic Class |
|---|---|
| Subject Ethnicity | Ethnicity |
| Subject Race | |
| Maximum Age | Age |
| Minimum Age | |
| Gestational Age | |
| Subject Gender | Gender |
| Pregnancy | Pregnancy-related Activity |
| Nursing | |
| Current Population Disease Condition; Past Population Disease Condition |
Disease, Symptom and Sign |
| Disease Stage | |
| Neoplasm Status | |
| Prior and Concomitant Medication | Pharmaceutical Substance or Drug |
| Substance Use | |
| Special Population | Special Patient Characteristic |
| Ethical Consideration | Consent |
| Compliance with Protocol | |
| Capacity | |
| Patient Preference | |
| Enrollment in Other Studies | |
| Lifestyle Choices | Bedtime |
| Diet | |
| Exercise | |
| Addictive Behavior | |
| BMI | Diagnostic or Lab Results |
| Address | |
| Allergy | |
| Device | |
| Life Expectancy | |
| Literacy | |
| Organ or Tissue Status | |
| Receptor Status | |
| Therapy or Surgery |
We observed that some of the highly prevalent eligibility criteria classes were not defined in BRIDG model, such as “Therapy or Surgery”, which has 48% prevalence in our eligibility criteria corpus. About 64% clinical studies include criteria for class “Diagnostic or Lab Results”, but the BRIDGE model only defined a very specific data item that maps to this class, which was BMI. One implication of this comparison study is that corpus-based knowledge acquisition or domain modeling can be as good as human experts and is promising to complement domain expertise and achieve more complete coverage of semantic classes.
DISCUSSION
Our rationale to identify semantic classes of eligibility criteria is not only to facilitate knowledge representation of eligibility criteria, but also to support natural language processing of eligibility criteria in two ways. First, different types of biomedical texts often require grammar rules for parsing needs; development of such rules is often laborious and time-consuming. One of our future tasks is to analyze the semantic patterns and define grammar rules for each induced eligibility criteria class to achieve a sublanguage of clinical research eligibility criteria. Knowledge of semantic classes can help us more accurately define class-specific information structure and grammar rules. Second, automatic categorization of an eligibility criterion can facilitate automatic selection of a specialized parser or grammar rules during natural language processing.
As a pilot study, our method has a couple of limitations. First, the manual process for cluster grouping and merging and class naming may contain inherent bias. Second, complex eligibility criteria often have several atomic criteria in one sentence, and hence do not necessarily map to a single category. We need to either extend our classifier to detect multiple classes in one sentence or automatically rewrite complex criteria into logical combinations of simple criteria before classification. Third, the UMLS semantic types might not provide the ideal granularity required by some decision support applications. Future work is needed to improve the manual effort for interpreting and adjusting clustering results or to evaluate the suitability of the classes for different applications.
CONCLUSION
We contribute a novel approach to corpus-based knowledge acquisition with two key designs: (1) using an UMLS-based semantic annotator to reduce the semantic features space and to remove semantic ambiguities, which are the common roadblocks to the traditional “bag of words” feature representation method; (2) using a hierarchical clustering method to reduce manual efforts required to identify semantic classes from text with minimal user input. Our results are comparable to that developed by domain expertise (e.g., BRIDGE). This approach is promising and worth further extension.
Acknowledgments
This research was funded under NLM grant R01 LM009886 and CTSA award UL1 RR024156. The authors thank the reviewers for their valuable feedback to an earlier version of this paper. We thank Yalini Senathirajah for her help with using the Hierarchical Clustering software. We also thank Drs. Herbert S. Chase, Lorena Carlo, and Meir Florenz for manually categorizing the eligibility criteria corpus.
REFERENCES
- 1.Weng C, Tu SW, Sim I, Richesson R. Formal representation of Eligibility Criteria: A Literature Review. Journal of Biomedical Informatics. 2010 doi: 10.1016/j.jbi.2009.12.004. (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lin D, Pantel P. San Francisco, California: 2001. Induction of semantic classes from natural language text. Paper presented at: KDD ‘01: Proceedings of the seventh ACM SIGKDD international conference on Knowledge discovery and data mining; pp. 317–322. [Google Scholar]
- 3.Pantel P, Ravichandran D. Human Language Technology Conference of the North American Chapter of the Association for Computational Linguistics. Boston, Massachusetts: 2004. Automatically Labeling Semantic Classes. [Google Scholar]
- 4.Niu Y, Hirst G. Analysis of Semantic Classes in Medical Text for Question Answering. Proceedings of the 7th Meeting of the ACL Special Interest Group in Computational Phonology: Current Themes in Computational Phonology and Morphology. 2004 [Google Scholar]
- 5.Cheng J, Cline M, Martin J, Finkelstein D, Awad T, Kulp D, Siani-Rose M. A Knowledge-Based Clustering Algorithm Driven by Gene Ontology. Journal of Biopharmaceutical Statistics. 2004;14(3):687–700. doi: 10.1081/bip-200025659. [DOI] [PubMed] [Google Scholar]
- 6.Pratt W, Fagan L. The Usefulness of Dynamically Categorizing Search Results. Journal of the American Medical Informatics Association. 2000 Nov;7(6):605–617. doi: 10.1136/jamia.2000.0070605. 2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lin D. Automatic Retrieval and Clustering of Similar Words. Paper presented at: COLING-ACL1998. :768–774. [Google Scholar]
- 8.Lewis DD. Naive (Bayes) at Forty: The Independence Assumption in Information Retrieval. In the Proceedings of ECML-98, 10th European Conference on Machine Learning. 1998:4–15. [Google Scholar]
- 9.Luo Z, Duffy R, Johnson S, Weng C. AMIA Summit on Clinical Research Informatics. San Francisco, California: 2010. Corpus-based Approach to Creating a Semantic Lexicon for Clinical Research Eligibility Criteria from UMLS; pp. 26–30. [PMC free article] [PubMed] [Google Scholar]
- 10.McCray AT. Better Access to Information about Clinical Trials. Annals of Internal Medicine. 2000;133(8):609–614. doi: 10.7326/0003-4819-133-8-200010170-00013. [DOI] [PubMed] [Google Scholar]
- 11.Seo J, Shneiderman B. A Rank-by-Feature Framework for Interactive Exploration of Multidimensional Data. Information Visualization. 2005;4(2):99–113. [Google Scholar]
- 12.Fridsma DB, Evans J, Hastak S, Mead CN. The BRIDG Project: A Technical Report. Journal of the American Medical Informatics Association. 15(2):130–137. doi: 10.1197/jamia.M2556. [DOI] [PMC free article] [PubMed] [Google Scholar]