Abstract
Post-marketing pharmacovigilance is important for public health, as many Adverse Drug Events (ADEs) are unknown when those drugs were approved for marketing. However, due to the large number of reported drugs and drug combinations, detecting ADE signals by mining these reports is becoming a challenging task in terms of computational complexity. Recently, a parallel programming model, MapReduce has been introduced by Google to support large-scale data intensive applications. In this study, we proposed a MapReduce-based algorithm, for common ADE detection approach, Proportional Reporting Ratio (PRR), and tested it in mining spontaneous ADE reports from FDA. The purpose is to investigate the possibility of using MapReduce principle to speed up biomedical data mining tasks using this pharmacovigilance case as one specific example. The results demonstrated that MapReduce programming model could improve the performance of common signal detection algorithm for pharmacovigilance in a distributed computation environment at approximately liner speedup rates.
Introduction
Adverse Drug Events (ADEs) are serious problems for public health. Adverse drug reactions are associated with 10.7% of hospital admissions in older adults (1, 2). Every year, ADEs cause more than 100,000 deaths in the United States, making ADEs the fourth leading cause of death in the United States (3). However, many ADEs are not known when those drugs were approved for marketing by FDA, because premarketing studies are usually limited in generalizability due to their limited size, short duration, and exclusively selected patient groups. For example, in 2004, five years after FDA’s approval in 1999, Merck & Co., Inc. withdrew Vioxx (Rofecoxib) from the market due to associated increased risk of heart attack and stroke (4, 5).
Therefore, post-marketing pharmacovigilance has become increasingly important. Over recent years, collecting ADEs electronically and detecting signals of unknown drug side effects using data mining approach have become popular (6). For example, FDA has established an on-line voluntary reporting system, the Adverse Event Reporting System (AERS). This system is designed to support the FDA’s post-marketing safety surveillance for all approved drugs. Among the data mining algorithms, the mostly used is the measures of disproportionality (6). Calculations of measures of disproportionality are primarily based upon a two-by-two contingency table (see Table 1).
Table 1.
The contingency table for ADE detection
| Target AE(s) | Other AE(s) | |
|---|---|---|
| Target Drug(s) | A | B |
| Other Drug(s) | C | D |
The commonly used measures for detecting ADEs based on Table 1 are summarized in Table 2.
Table 2.
Summary of measures of disproportionality
| Definition | |
|---|---|
| Reporting Odds Ratio (ROR) | |
| Proportional reporting ratio (PRR) | |
| Yules Q ratio |
However, due to the large volume and rapid growth of ADE reports and the large number of reported drugs and adverse effects, mining these reports is becoming a challenging task in terms of computational complexity. For example, in the ADE reports collect from FDA, the number of reported drugs and adverse effects are 237,579 and 14,401 respectively. Theoretically, we have 3,421,375,179 number of 1-drug and 1-effect combinations. This number will increase exponentially if the combinations of arbitrary number of drugs and effects are considered. Different from previous studies which targeted on a limited number of selected drugs, our work on mining ADE reports is to evaluate all combinations for any existing drugs and effects. This becomes a computational intensive task, and practically is often beyond the capacity of a single computer node in terms of CPU and storage. Therefore, the distributed high performance computation model is necessary for this task.
Recently, MapReduce programming model has been introduced by Google to support large-scale data intensive applications, such as large-scale indexing for search engine (7). The typical MapReduce systems are Google MapReduce (7), Microsoft Dryad (8) and Apache Hadoop (9). In general, MapReduce includes two steps: map and reduce. In the map phase, a given problem is divided into smaller sub-problems where each sub-problem can be solved independently to others. In the reduce phase, the answers of all sub-problems are combined in a way to get the output which is the final answer of the original problem. Note that the results of the map phase are partitioned and all map results from different map nodes (mapper) but with the same key are assigned to the same reduce node (reducer). The advantage of MapReduce is that it exploits parallelism from low-cost workstation clusters to process both map and reduce operations and thus achieve comparable performance to high-end servers. Comparing with the centralized systems, MapReduce offers far better scalability and reliability at an affordable cost.
Although MapReduce has been widely used for massive data analytics, very little information exists on the application of MapReduce in the biomedical domain. In this study, we designed and implemented an association rule algorithm using the MapReduce paradigm and demonstrated how our approach can help in mining the spontaneous ADE reports from FDA. Therefore, the purpose of this paper is to investigate the possibility of using the MapReduce principle to speed up the process of detecting the ADE signals for pharmacovigilance and to introduce the MapReduce paradigm to the machine learning community in biomedicine.
Methods
The data for ADE analysis are from FDA, including the AERS reports collected from 1st quarter, 2004 to 4th quarter, 2009, totally 22 data files. The drug names in the reports can be classified into two types: accurate name and report name. While the former is the standard name with confirmation, the latter is the original name input by reporter, which could be inaccurate. In the ADE reports, there are 5,694 types of accurate drug names, 233,242 types of report drug names, and 14,401 types of effect names. Considering data quality, we generate two datasets: AERS-1 consists of the reports that only contain accurate drug name, and AERS-2 is the complete set of reports. Table 3 lists the features of these datasets.
Table 3:
Features of AERS datasets.
| AERS-1 | AERS-2 | |
|---|---|---|
| Number of reports | 1,236,951 | 1,963,588 |
| Total number of drug items | 2,411,984 | 6,720,986 |
| Total number of effect items | 4,091,686 | 7,487,581 |
| Avg. number of drug items | 1.95 | 3.42 |
| Avg. number of effect items | 3.30 | 3.81 |
| Avg. number of items per report | 5.25 | 7.24 |
| Size | 46M | 109M |
With the purpose of identifying the possible associations between drugs and adverse effects, we take the ADE analysis task as a risk pattern discovery problem. A risk pattern is in the form of (DrugSet, EffectSet), where DrugSet and EffectSet are the set of drug(s) and adverse effect(s) respectively.
Similar as the work (12), besides applying PRR≥2 and Chi-square≥4 as the measures to evaluate the significance of the risk pattern, we adopt the important condition called minimum count. The minimum count condition requires that the number of ADE reports that contain the drugs and effects, denoted as N(DrugSet ∪ EffectSet), is no less than 3 (set by domain expert). This condition is imposed for excluding the patterns with extremely rare occurrences, because they could be unconvinced as signals statistically.
Thanks to minimum count condition, our MapReduce approach can be divided into two tasks.
Task 1 finds all sets of drugs, effects, or drug-effect combinations that occur in at least 3 ADE reports. We call these sets of items frequent itemsets. The output of Task 1 is all frequent itemsets with their count, denoted as {<itemset, N(itemset)>}. Apparently, the itemset could be DrugSet, EffectSet, or DrugSet ∪ EffectSet. Task 1 is the key to ADE analysis because it not only extracts the candidate risk patterns by eliminating the large number of invalid drug-effect combinations, but also gets the counts for computing PRR and Chi-Square.
Task 2 computes the PRR and Chi-square for the all candidate risk patterns and output significant ones.
Task 1: Finding frequent itemsets. Finding frequent itemset is an important task of data mining. Apriori approach (10) is a classic solution for this problem. However, it does not work for the ADE analysis due to the large number of drug-effect combinations and the low minimum count threshold. Therefore, we proposed and implemented a MapReduce-based approach, DistApriori, on Hadoop platform (9).
DistApriori follows the basic steps of Apriori. The key property utilized by this approach is that any subset of a frequent itemset must be frequent. For example, if a 3-itemset {1, 2, 3} is frequent, all of its sub 2-itemsets {1, 2}, {1, 3}, and {2, 3} must be frequent. According to this property, DistApriori firstly finds the frequent 1-itemsets by scanning the ADE reports. Then it iteratively uses k-itemset (k is the length of itemset and k≥1) to generate candidate (k+1)-itemsets, and scans the data to find the frequent (k+1)-itemsets. This iteration stops until no candidate (k+1)-itemset can be generated. Compared with Apriori, the key difference of DistApriori is that in each iteration, the generated candidate itemsets are partitioned so that each computer in the cluster can effectively process a fragment of them.
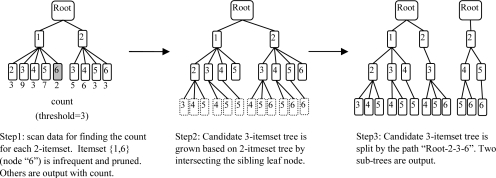
Candidate itemset tree: In order to effectively partition the candidate itemsets, we organize the candidate itemsets as a lexicographic ordered tree. Each node in a candidate itemset tree is an item id. For a candidate k-itemset tree, each path from root to leaf forms a k-candidate pattern. The nodes on the tree are ordered numerically according to item id, i.e., the parent node is smaller than child node and the sibling node on the left is smaller than that on the right. For example, in Figure 1, the left tree is a candidate 2-itemset tree and leaf node “2” represents a candidate itemset {1,2}.
Figure 1:
Example of map phase in Task 1
When generating the candidate itemset tree, if the tree becomes too large for a single computer, DistApriori will split the tree and assign the sub-trees to different computer nodes. This process can be implemented as a map phase only (i.e., no reduce phase required). Figure 1 illustrates the three steps of the map phase in an iteration.
A candidate k-itemset tree is loaded and ADE reports are scanned to find the count for each candidate k-itemset. Frequent k-itemsets are output, and infrequent ones are pruned from the tree.
Candidate (k+1)-itemset tree are generated by intersecting leaf items with the same parent node.
Candidate (k+1)-itemset tree is split to ensure each sub-tree could be fit in memory for processing. And the sub-trees are output to distributed file system as the input of next round.
Task 2: Computing PPR and Chi-Square for risk patterns. For each frequent itemset including both drugs and effects (DrugSet ∪ EffectSet), we need to find the count of DrugSet and EffectSet. This is not an easy task because we are processing millions or even billions of itemsets. So we design a two-round MapReduce process for Task 2.
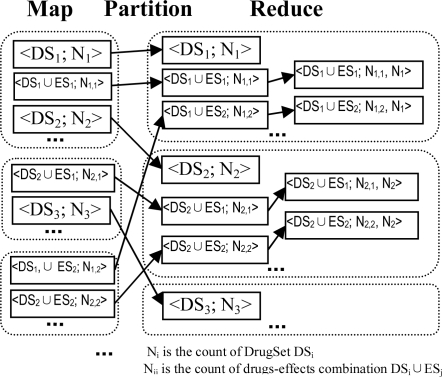
First round MapReduce. In this round, we aggregate all data entries by drugs to get the count of drug set for each drug-effect entry. The main process is depicted in Figure 2. In map phase, we simply output each entry directly. Then, we design the partition such that all entries containing the same drug set DSi are sent to the same reducer, including the entry of DSi itself. In reduce phase, for the drug set DSi, we get its count Ni and append it to drug-effect entries that contain the drug set DSi.
Second round MapReduce. In this round, we aggregate entries (output of 1st round) by effects using the similar process as the first round, and get the count of effect set for each drug-effect entry. In reduce phase, after getting the values for A, B, C and D in Table 1, we calculate PRR and Chi-square for each drug-effect entry. Finally, the drug-effect entries which meet the criteria are output as risk patterns.
Figure 2:
MapReduce process of Task 2
Results
We conduct our studies on top of a Hadoop cluster, which has 20 computer nodes interconnected by 1GB Ethernet. Each node is a Dell server with Intel Xeon 2.8GHz CPU (4 cores inside), 4GB RAM and 1TB 7200RPM hard driver. The operating system of all nodes is Red Hat Enterprise Linux AS 4 (RHEL4) and the version of the Hadoop platform is 0.20.1.
Although we applied the MapReduce approach, the required data storage for AERS analysis is beyond of the capacity of our cloud cluster. To control the data size, mainly the number of candidate itemsets, we add a constraint on the length of ADE reports based on preliminary experiment results. Any report whose item length is greater than a threshold will be ignored.
The analysis result is shown in Table 4. We set the length threshold of ADE reports as 16 and 20 respectively. For example, for AERS-1 dataset, we get 96.7% (1196078) of all records when the threshold is16. In each experiment, a large number of risk patterns are discovered and ordered by PRR. Note that with the increase of minimum count threshold (now it is 3), the number of risk patterns can be significantly reduced.
Table 4:
Analysis result
| Dataset | Length threshold | # report (percentage) | # frequent itemsets | # risk patterns |
|---|---|---|---|---|
| AERS-1 | 16 | 1,196,078 (96.7%) | 25,385,057 | 18,697,485 |
| AERS-1 | 20 | 1,213,531 (98.11%) | 160,422,115 | 124,047,300 |
| AERS-2 | 16 | 1,799,862 (91.66%) | 84,503,728 | 65,830,916 |
| AERS-2 | 20 | 1,867,855 (95.12%) | 573,712,809 | 476,610,821 |
Scalability We evaluate the scalability of our approach in terms of the number of nodes in the cluster. For four different dataset settings, we conduct the tests on the clusters with 5, 10, 15, and 20 nodes respectively. Table 5 gives the execution time for Task 1, Task 2, and Total for each test.
Table 5:
Execution time (minute)
| Dataset and settings | # Nodes | 5 | 10 | 15 | 20 |
|---|---|---|---|---|---|
| AERS-1-16 | Task 1 | 59.53 | 32.12 | 26.12 | 20.17 |
| Task 2 | 27.26 | 14.12 | 10.18 | 7.88 | |
| Total | 86.79 | 46.24 | 36.30 | 28.06 | |
| AERS-1-20 | Task 1 | 97.21 | 49.25 | 38.39 | 29.63 |
| Task 2 | 83.80 | 44.45 | 34.56 | 22.97 | |
| Total | 181.01 | 93.71 | 72.94 | 52.60 | |
| AERS-2-16 | Task 1 | 223.26 | 111.48 | 79.96 | 58.63 |
| Task 2 | 65.04 | 34.06 | 23.61 | 17.41 | |
| Total | 288.30 | 145.53 | 103.57 | 76.04 | |
| AERS-2-20 | Task 1 | 439.99 | 230.05 | 156.00 | 114.23 |
| Task 2 | 274.70 | 143.99 | 102.07 | 85.48 | |
| Total | 714.70 | 374.03 | 258.07 | 199.71 |
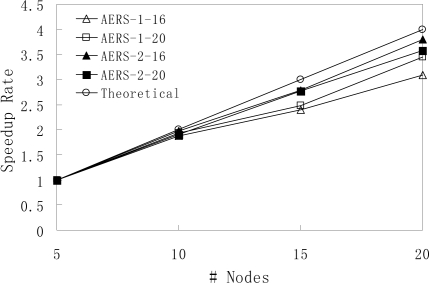
Figure 3 depicts the speedup rate of our approach in terms of the number of nodes in the cluster. We can see that for all four dataset settings, our MapReduce-based approach reaches the linear scalability. Importantly, it means that we can improve the performance proportionally when more computers are added into the cluster.
Figure 3:
Scalability Evaluation
Comparison To compare the performance of cloud-based approach and the centralized approach, we implement a serialized algorithm1 for ADE detection. We run the algorithm on a single computer for the dataset AERS-1 and AERS-2 dataset, with the length threshold as 20. The results of running time are 338.32 minutes and 1505.47 minutes respectively. Compared with the corresponding results from the cloud with 5 nodes (181.01 minutes and 714.7 minutes), the ratios are 1.87 and 2.10 respectively. It seems that the cloud-based approach does not explore the full computation power of each node. However, this is reasonable because in the cloud, there will be the additional cost for parallel computation, such as network I/O, backup I/O, and sorting process. So, we still can be confident that MapReduce is effective for improving the performance of data processing, especially when the computation over a larger dataset is improbable in a single machine.
A preliminary manual evaluation has been conducted on the first 100 patterns obtained from AERS-2 dataset with the highest Chi square values. Interestingly, we found that most of the 100 patterns are about chemotherapies which contain two to five chemotherapeutic drugs, including dexamethasone acetate, Oncovin, Alkeran, Endoxan, and doxorubicin hydrochloride. This may demonstrate that chemotherapeutic drugs can lead to more adverse events than other drugs. The major detected adverse events include instillation site pruritus, a common effect in patients with long-term instillation, and accessory nerve disorder, a possible result of peripheral neuropathy.
Discussion
The results have successfully demonstrated that MapReduce programming model could be used to improve the performance of common signal detection algorithm for pharmacovigilance in a distributed computation environment. Our proposed approach has the following advantages over single computer platforms. First, the MapReduce approach provides a solution to the computation-intensive task of pharmacovigilance that could be very difficult or even impossible for a single regular PC server. Second, the proposed approach has a very good extensibility. More computational capability could be easily acquired by adding more workstations to the networked environment. The result of our experiment on a 20-node environment has shown pretty liner speedup rates. Third, because all the workstations can use ordinary x86 PCs, this provides an affordable and easy solution for high-performance computation. For example, the computation environment could even be setup in a regular computer class-room of a university. Forth, the environment has innate fault-tolerance since the task of a node could be transferred to other nodes when the workstation at the node experiences a hardware failure.
Our study demonstrated MapReduce methodology for biomedical data mining task by one specific example in detecting ADEs for pharmacovigilance. We are aware of the following limitations in this study. First, we conducted the experiment on a cluster platform of only 20 nodes. Although the results have shown approximately linear speedup rates, whether the linear speedup can still be maintained in hundred or thousands nodes is still unconfirmed. Furthermore, rewriting regular mining algorithm according to MapReduce model needs additional programming work. This may affect the general use of MapReduce in biomedical domain. Additionally, MapReduce model has its own limitations in that it requires the task is dividable into small sub-tasks so that they can be processed at different nodes. Some studies have reported that the characteristics of different machine learning algorithm may lead to different speedup rates (11).
Although applied for detecting ADEs for pharmacovigilance, we believe that the use of MapReduce paradigm can be extended to solve other data mining tasks in biomedical domain. First, some experiments have corroborated that MapReduce can speed up a variety of common machine learning algorithms, such as naïve Bayes, k-means, logistic regression, neural network, principal components analysis, support vector machine, and so on (11). On the other hand, the rapidly growing electronic data in biomedicine, such as electronic medical record, will provide the rich material and topics for data mining explorations. We hope that this report on a specific example could arouse the general interest of biomedical data mining community in this new parallel programming model, MapReduce.
Conclusion
As an early study of using MapReduce approach in biomedical data mining task, we demonstrated that MapReduce programming model could improve the performance of common signal detection algorithm for pharmacovigilance in a distributed computation environment at an approximately liner speedup rate. MapReduce paradigm represents a promising direction for computation-intensive biomedical tasks.
Footnotes
The serialized algorithm follows the basic steps and the partitioning methods in our MapReduce approach, but processes the data partitions in serial rather than in parallel.
References
- 1.Moore TJ. Prescription for disaster: the Hidden Dangers in Your Medicine Cabinet. New York, NY: Simon & Schuster; 1998. [Google Scholar]
- 2.Kongkaew C, Noyce PR, Ashcroft DM. Hospital admissions associated with adverse drug reactions: a systematic review of prospective observational studies. Ann Pharmacother. 2008 Jul;42(7):1017–25. doi: 10.1345/aph.1L037. [DOI] [PubMed] [Google Scholar]
- 3.Lazarou J, Pomeranz BH, Corey PN. Incidence of adverse drug reactions in hospitalized patients: a meta-analysis of prospective studies. Jama. 1998 Apr 15;279(15):1200–5. doi: 10.1001/jama.279.15.1200. [DOI] [PubMed] [Google Scholar]
- 4.Mukherjee D, Nissen SE, Topol EJ. Risk of cardiovascular events associated with selective COX-2 inhibitors. Jama. 2001 Aug 22–29;286(8):954–9. doi: 10.1001/jama.286.8.954. [DOI] [PubMed] [Google Scholar]
- 5.Bombardier C, Laine L, Reicin A, Shapiro D, Burgos-Vargas R, Davis B, et al. Comparison of upper gastrointestinal toxicity of rofecoxib and naproxen in patients with rheumatoid arthritis. VIGOR Study Group N Engl J Med. 2000 Nov 23;343(21):1520–8. doi: 10.1056/NEJM200011233432103. [DOI] [PubMed] [Google Scholar]
- 6.Wilson AM, Thabane L, Holbrook A. Application of data mining techniques in pharmacovigilance. Br J Clin Pharmacol. 2004 Feb;57(2):127–34. doi: 10.1046/j.1365-2125.2003.01968.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dean J, Ghemawat S. MapReduce: simplified data processing on large clusters. Proc of the 6th Symposium on Operating Systems Design & Implementation (OSDI’04); 2004. p. 2004. [Google Scholar]
- 8.Isard M, Budiu M, Yu Y, Birrell A, Fetterly D. Dryad: distributed data-parallel programs from sequential building blocks. SIGOPS. 2007;41(3) [Google Scholar]
- 9.Apache Hadoop http://hadoop.apache.org.
- 10.Agrawal R, Srikant R. Fast Algorithms for Mining Association Rules. Int Conf Very Large Data Bases; 1994. p. 1994. [Google Scholar]
- 11.Chu C-T, Kim SK, Lin Y-A, Yu Y, Bradski G, Ng A, et al. Map-Reduce for Machine Learning on Multicore. NIPS. 2006:2006. [Google Scholar]
- 12.Evans SJ, Waller PC, Davis S. Use of proportional reporting ratios (PRRs) for signal generation from spontaneous adverse drug reaction reports. Pharmacoepidemiol Drug Saf. 2001 Oct–Nov;10(6):483–6. doi: 10.1002/pds.677. [DOI] [PubMed] [Google Scholar]