Abstract
Clinical reporting is often performed with minimal consideration for secondary computational analysis of concepts. This fact makes the comparison of patients challenging as records lack a representation in a space where their similarity may be judged quantitatively. We present a method by which the entirety of a patient’s clinical records may be compared using latent topics. To capture topics at a clinically relevant level, patient reports are partitioned based on their type, allowing for a more granular characterization of topics. The resulting probabilistic patient topic representations are directly comparable to one another using distance measures. To navigate a collection of patient records we have developed a workstation that allows users to weight different report types and displays succinct summarizations of why two patients are deemed similar, tailoring and expediting searches. Results show the system is able to capture clinically significant topics that can be used for case-based retrieval.
Introduction
Case-based retrieval as a problem solving technique is a growing area of study and in medicine is frequently implemented by comparing quantitative values, such as blood glucose level or respiratory rate, across patients [1–2]. The similarity between a query patient and a test patient or reference standard (e.g., a hyperglycemia profile) is then determined by the differences between these values, adjusting for time and other factors that characterize the targeted feature of interest. Although this approach is effective for certain types of retrieval and monitoring, it is limited in that it: 1) requires computer interpretable structure in the medical record upon which to directly compare and reason, which may not be available; and 2) ignores the totality of information stored in a patient’s free-text medical record.
Clinicians at our institution maintain data repositories of patients with particular diseases, such as brain cancer, for secondary purposes that include research (e.g., cohort identification) and education (e.g., teaching files), as well as assisting in clinical diagnosis, treatment and prognosis. The last set of tasks is performed by comparing data from a new patient of interest with data from patients with known outcomes. In general, these clinician-maintained repositories are rudimentary lists of patient IDs pointing to medical records, as well as additional data stored in a local database specific to the clinicians’ research (e.g., specific quantitative data or observations that may not be clinically reported). To gain a broader view of a patient, many clinicians would like to augment the data they maintain within their practice with information from other sources in the hospital. However, given the current disparate and unstructured electronic reporting infrastructure (e.g., both radiology and pathology maintain separate databases with non-standard schemas), it is prohibitively expensive and time-consuming for one to manually retrieve, process, and extract this information. To help overcome these issues we have developed a workstation that automatically structures patient records with latent topics, providing a mechanism for search and case-based retrieval.
Methodology
Document Collection
We investigated the use of topic models for case-based retrieval in a population of patients with glioblastoma multiforme (GBM), an aggressive brain cancer [3]. Our initial corpus contained 324 patients. Each patient’s medical record was partitioned into bins based on report type. These types were selected by a clinician to capture critical sources of information for a patient with GBM and are listed in Table 1. The binning process was performed using meta-information included within a report in our hospital information system as well as by matching known regular expressions within a report’s title.
Table 1.
Summary of data set detailing the report types used, the number of reports, the number of unique tokens pre- and post-processing and the number of topics, K, in each LDA model.
| Report Types | # Reports | # Unique Tokens | K | |
|---|---|---|---|---|
| Pre | Post | |||
| MRI Brain | 3,409 | 31,209 | 3,028 | 50 |
| CT Brain | 371 | 7,575 | 1,556 | 30 |
| PET Brain | 105 | 3,397 | 813 | 40 |
| Other Radiology | 1,247 | 22,888 | 4,483 | 50 |
| Oncology Progress | 3,370 | 67,773 | 6,787 | 40 |
| Neurosurgery Operative | 414 | 16,640 | 3,964 | 35 |
| Discharge Summary | 553 | 26,212 | 5,539 | 40 |
| Pathology | 117 | 6,268 | 1,685 | 50 |
Document Model
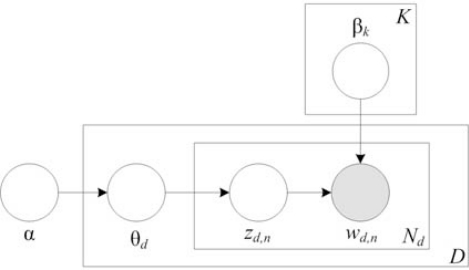
Underpinning the retrieval system, documents are characterized in a topic space using latent Dirichlet allocation (LDA) [4]. Latent variable models, such as LDA, assume that within a collection of text there exists a set of semantic topics expressed by patterns of word usage across a corpus. Therefore, a document may be modeled as a mixture of these topics, which are then responsible for generating words. Fig. 1 provides a graphical model depiction of LDA, which illustrates the conditional dependencies between random variables [5]. Shaded nodes represent observed variables, while boxes represent replication, i.e., a corpus contains D reports and a report contains Nd words. In LDA, a multinomial sample, θ, is drawn from a Dirichlet distribution for each document, d, and specifies a distribution over K topics. The multinomial document-topic distribution is then sampled once for every word n in a report to select a topic, z, which indexes the word-topic distribution, β, from which words are drawn. The Dirichlet parameter, α, and the multinomial topic distributions, β, may be learned with standard parameter estimation techniques. In this work, the MALLET toolkit, which uses Gibbs sampling, was used to fit the model to the data [6].
Figure 1.
Graphical model of latent Dirichlet allocation (LDA). Boxes denote replication.
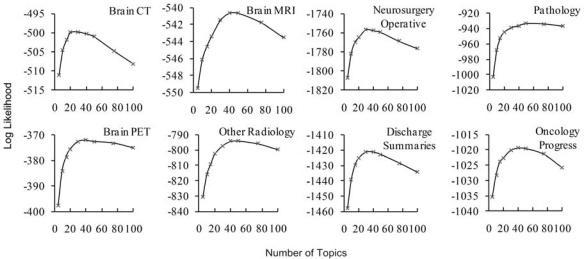
An LDA model was created for each selected report type, allowing for granular topics to be learned, rather than more general topics across all types (which arguably would provide a clinically less specific means of comparison). To estimate the number of topics in each LDA model, 20% of the data was randomly selected and held-out for testing. The trained model was then used to infer document topic distributions on the test data and the log likelihood (a goodness-of-fit measure) was computed. The number of topics used in each model was selected at the point after which log likelihood decreased (see Fig. 2).
Figure 2.
Test set log likelihood for LDA models.
Document Pre-processing
Before fitting the LDA model, documents are tokenized by whitespace and pre-processed to remove stop-words, punctuation, numbers, symbols, and any patient- or physician-identifiable information. Next, because LDA seeks differences in word patterns across documents and the language used in the reports of GBM patients is much less varied in comparison to the areas where LDA has been applied in the literature (e.g., newspaper articles), any word appearing in over 75% of the reports was discarded. For example, within brain MRI reports the word “tumor” appeared 90% of the time, giving it little discriminating power. One may pursue phrasal analysis to distill a wider variety of clinical concepts; however, our results show that even by using unigrams we are able to discover clinically relevant topics and require less data to do so. Finally, to further distill clinically-relevant concepts, the remaining tokens are compared to terms in the Unified Medical Language System (UMLS). Any token that does not match an STR entry in the MRCONSO table is discarded. Table 1 provides an overview of the dataset, including the number of unique words pre- and post-processing.
Similarity Metric and Retrieval Algorithm
Under LDA, a document is ultimately represented as a distribution over latent topics. To make comparisons across patients, document topic distributions were summed and normalized, i.e., for each bin a multinomial distribution over topics, pbin is computed where each element, k, of pbin is calculated as:
With a topic representation of each patient in hand (one for each type of report), comparisons between patients can be made by computing the symmetric Kullback-Leibler divergence between two patient-topic distributions for a given report type, b. These divergences may then be summed over all types to provide a measure of how similar two patients are in total (i.e., smaller sums indicate similarity). In addition, a set of user-defined weights, ω, allows one to emphasize different report types in a search. Thus, given a collection with type bins {1,...,b,...,B} having Kb topics per bin, for query patient q, the distance between any other patient, p, in the collection is defined as:
For a given query patient, the distance, Dqp, between each patient in the collection may be calculated and returned in sorted order to the user, thereby enabling similarity retrieval based on this metric.
Results and Discussion
For each model, Fig. 2 shows the number of topics vs. the test set log likelihood; a measure of the probability that the trained model generated the test data. As more topics are added, patterns of words are better fit by the model. This trend changes at the point where adding additional topics acts only to capture the variation specific to the training set and likelihood of the test set decreases. Table 1 specifies the number of topics used for each report type. Table 2 shows example topics from each report type and suggested labels.
Table 2.
Example topics and their suggested labels for each report type. The six most probable terms are shown.
| Topic | Label | |
|---|---|---|
| MRI | edema, midline, shift, vasogenic, large, ventricle | Edema and midline shift |
| blood, products, consistent, amount, margins, degradation | Resection cavity status | |
| CT | maxillary, thickening, mucosal, sinus, retention, cyst | Inflammation/infiltrate within sinus cavity |
| enhanced, irregular, bleeding, shows, abnormality, contrast | Stroke/vasospasm | |
| PET | occipital, edema, surrounding, suggestive, malignancy, cortex | Tumor assessment |
| parietal, demonstrated, previous, mid, glucose, hemisphere | Comparison glucose uptake | |
| Other Rad. | pulmonary, lower, lobe, chest, angiogram, atelectasis | Chest x-ray evaluation |
| patient, pain, back, compression, vetebroplasty, fractures | Spinal MR assessment for back pain | |
| Neuro | lesion, biopsy, center, stereotactic, bleeding, frozen | Surgical resection and biopsy |
| catheter, shunt, ventricular, peritoneal, valve, hydrocephalus | Shunt placement during resection | |
| Discharge | assistance, functional, activities, required, mobility, living | Quality of life assessment |
| difficulty, speech, grade, obtained, glioma, frontotemporal | Neurological functional assessment | |
| Pathology | mass, excision, calcification, special, pink, intensity | Tumor histopathology and staining |
| mitoses, hyperplasia, infiltrating, estimated, focally, cellularity | Malignant biopsy | |
| Oncology | difficulty, memory, shows, short, term, slight | Decreased neurological function |
| accutane, days, cycle, completed, frontal, follow | Chemotherapy plan |
Retrieval Workstation
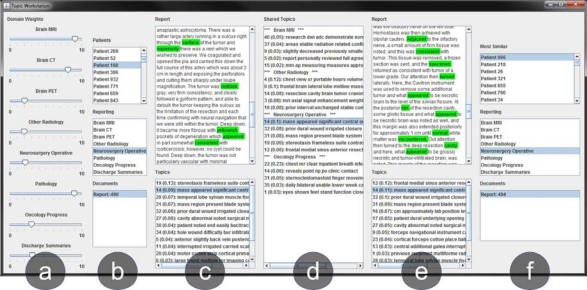
Detailed in Fig. 3 is a retrieval workstation built using Java Swing. The goal of the interface was to provide a mechanism by which clinicians could navigate patient records and execute case-based retrieval queries. Users may view a patient’s reports by type and, for each report, view the most prevalent (latent) topics within it. When a patient is selected, the results list of patients is sorted by distance to the given selection. Because LDA models distributions of topics in documents, when a given report from a query patient is selected, the reports for the resulting patients may be sorted based on KL divergence (i.e., if a user selects a brain MRI report from a query patient, all the brain MRI reports from the result patient will be sorted). When viewing a report and a topic is selected, words generated by that topic are highlighted, allowing the user to quickly see where the meaning of the topic is conveyed.
Figure 3.
Topic case-based retrieval workstation. (a) Adjustable weights for reporting domains. (b) Query patient information. (c) Highlighted reports and topics for query patient. (d) Clickable shared topics between patients. (e) Highlighted result reports and topics. (f) Most similar patients listed in descending order for selected query patient.
A user may also modify their query by modifying the weight for a report type using a set of slider bars. We found that clinicians were interested in weighting types differently depending on the relationships they were interested in. For example, a neuro radiologist was interested in assigning high weights to the brain MRI and pathology report types to retrieve tumors with similar imaging and histologic presentations.
Although two patients may have many topics in common, they typically have different numbers of reports and therefore different expressions of the common topics across their reports. Thus, a challenge we faced was how to succinctly show the clinician the ways in which two patients were most similar. Our approach was to calculate and display the topics most in common between patients, allowing the user to navigate records through the common topics. For two patients, the most common topic is judged to be the one with the largest expression in both cases (see Table 3). The list of shared topics is displayed between the query and the result. A user may click a shared topic and the reports for the query and result sort in descending order based upon the prevalence of the selected topic in a report. Words from the selected topic are also highlighted in the reports. Clinicians found this mechanism to be a more effective way of searching rather than sifting through each patient’s report in the results list to find matching topics to the query.
Table 3.
Two example patient topic distributions with the three most common topics highlighted.
| Topic Number | ||||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | |
| Patient 1 | 0.01 | 0.2 | 0.02 | 0.05 | 0.26 | 0.01 | 0.1 | 0.03 | 0.3 | 0.02 |
| Patient 2 | 0.04 | 0.14 | 0.1 | 0.01 | 0.2 | 0.01 | 0.01 | 0.04 | 0.4 | 0.03 |
Examples
Table 4 provides several examples of the types of relationships the system finds between patients. Shown are excerpts from the query patient’s record and similar text from the top retrieved patient record in the top matching document as inferred by the shared topic. The examples illustrate the ability of LDA to capture the variety of words and ways in which a clinical concept is conveyed, consolidating them into a single topic whose expression can be measured across patients as an indicator of similarity.
Table 4.
Similar concepts from retrieval results. The top six words from the topic shared between patients is presented along with the matching free-text. Words discarded in pre-processing are left in for clarity.
| Query Patient | Top Result | Topic |
|---|---|---|
| “...is a small amount of thick and nodular enhancement in the region of the left genu of the corpus callosum...” | “...minor contrast blushing in the left side of the corpus callosum...” | corpus, callosum, genu, splenium, midline, extending |
| “Her naming is impaired as well as her repetition. Her comprehension is fairly reasonable.” | “He is awake, alert, has minor difficultly with orientation and attention. His short-term short-term memory is impaired.” | difficulty, memory, shows, short, term, slight |
| “...status post right frontal glioblastoma resection...” | “...the patient has undergone a right frontal craniotomy for resection of the mass...” | frontal, medial, sinus, anterior, resected, medially |
| “He has normal language function and visual spatial skills... can stand with his feet together, eyes opened, eyes closed.” | “She has normal language function and visual spatial skills... could stand with her feet together, eyes open, eyes closed.” | eyes, shoes, feet, stand, function, closed |
| “The cells are arranged in sheets...with coarse granular chromatin. Larger cells show irregular nuclear borders.” | “The glial tumor is comprised of sheets... vesiculated nuclei with clumped chromatin, and irregular nuclear contours.” | chromatin, measure, normal, parts, pale, clusters |
| “The neoplasm extends along the ependymal surface of the ventricle.” | “...irregular ependymal enhancement involving the frontal horn of the right lateral ventricle.” | ventricle, horn, ependymal, spread, atrium, surface |
Conclusion
We have demonstrated the application of a topic model in discovering relevant clinical concepts and structuring a patient’s medical record. The imposed statistical structure was then used for case-based information retrieval of similar patients. The analysis of the system in terms of precision and recall is challenging due to the exhaustive requirements of generating a gold standard. However, the generation and release of such a set is a point of future work. We are augmenting this system with additional query mechanisms including demographics and lab values. Additionally, we are pursuing query templates consisting of different weightings of domains to answer pre-defined clinical questions. Phrasal discovery and analysis for improved topic learning is also underway.
Our approach can be quickly applied to any type of clinical document corpus as it requires no customization. However, for document corpora with relatively limited variation in words and grammar, a customized, knowledge-driven approach may also be appropriate. Such a system would likely take much longer to create, but could ultimately better capture clinical notions of similarity.
We plan to explore new applications for topic models in clinical reporting and have begun implementing techniques for 1) topic-driven problem list generation; and 2) systems that analyze the expression of a topic over time for modeling the progression of a disease process.
Acknowledgments
This research was funded by the National Institutes of Health (R01-LM009961, R01-EB009306).
References
- 1.Schmidt R, Montani S, Bellazzi R, Portinale L, Gierl L. Cased-based reasoning for medical knowledge-based systems. International Journal of Medical Informatics. 2001;64(2–3):355–67. doi: 10.1016/s1386-5056(01)00221-0. [DOI] [PubMed] [Google Scholar]
- 2.Aamodt A, Plaza E. Case-based reasoning. Proc MLnet Summer School on Machine Learning and Knowledge Acquisition. 1994:1–58. [Google Scholar]
- 3.Holland E. Glioblastoma multiforme: the terminator. National Acad Sciences. 2000:6242–4. doi: 10.1073/pnas.97.12.6242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blei D, Ng A, Jordan M. Latent Dirichlet allocation. Journal of Machine Learning Research. 2003;3(5):993–1022. [Google Scholar]
- 5.Jordan M, editor. Learning in Graphical Models. Cambridge: MIT Press; 1999. [Google Scholar]
- 6.McCallum AK. MALLET: A Machine Learning for Language Toolkit.: http://mallet.cs.umass.edu;2002.