Abstract
Chickpea ranks third among the food legume crops production in the world. However, the genomic resources available for chickpea are still very limited. In the present study, the transcriptome of chickpea was sequenced with short reads on Illumina Genome Analyzer platform. We have assessed the effect of sequence quality, various assembly parameters and assembly programs on the final assembly output. We assembled ∼107million high-quality trimmed reads using Velvet followed by Oases with optimal parameters into a non-redundant set of 53 409 transcripts (≥100 bp), representing about 28 Mb of unique transcriptome sequence. The average length of transcripts was 523 bp and N50 length of 900 bp with coverage of 25.7 rpkm (reads per kilobase per million). At the protein level, a total of 45 636 (85.5%) chickpea transcripts showed significant similarity with unigenes/predicted proteins from other legumes or sequenced plant genomes. Functional categorization revealed the conservation of genes involved in various biological processes in chickpea. In addition, we identified simple sequence repeat motifs in transcripts. The chickpea transcripts set generated here provides a resource for gene discovery and development of functional molecular markers. In addition, the strategy for de novo assembly of transcriptome data presented here will be helpful in other similar transcriptome studies.
Keywords: De novo assembly, chickpea, next generation sequencing, transcriptome, short read
1. Introduction
Chickpea (Cicer arietinum L.) is the third most consumed legume crop, which is cultivated in arid and semi-arid areas around the world.1 Chickpea is a self-pollinated, diploid (2n = 2x = 16) and annual plant with a moderate genome size of about 740 Mb. Despite growing demand and high-yield potential, chickpea productivity is very low. Several biotic such as Ascochyta blight, dry root rot, Fusarium wilt and pod borer, and abiotic such as drought, salinity and low temperature, constraints are major factors for lower chickpea production. Modern breeding technologies with biotechnological techniques are required to increase the productivity.2 Unfortunately, very limited genomic information is available for chickpea.
Various genomic tools have facilitated greatly the development of improved genotypes/varieties in several crop species.3,4 Although a few expressed sequence tags (ESTs) have been generated and gene-based markers have been developed, the functional genomics studies in chickpea is still in its infancy. Most of the ESTs have been generated with the aim to identify the candidate genes involved in various abiotic and biotic stress responses and development of molecular markers.5–9 In addition, other microarray and SAGE technologies have also been used to identify the stress-responsive transcriptome in chickpea.10,11 The efforts have been made to clone genes of interest via candidate gene approach.2 The function of a few genes in stress responses has also been demonstrated using transgenic approach.12,13
The generation of large-scale ESTs is a very useful approach to accelerate the research on non-model species. Although ESTs and other cDNA sequences are among the most reliable evidences for the identification of gene-rich regions in a genome, gene identification and genome annotation, very less effort has been made for chickpea in this direction when compared with other crop plants. This is reflected by a very small number of ESTs (34 587) present in the dbEST database at NCBI (release 100110; 1 October 2010) for chickpea. The next generation sequencing technologies provide a cost-effective means of sequencing the transcriptome of an organism.14 Several studies have reported the transcriptome sequencing of various model and non-model species using these technologies. However, most of these studies are based on the long-read sequence data using 454 pyrosequencing or employing hybrid approach.15,16 Although the efforts have been made to develop tools for de novo assembly using short-read sequence data,17–19 their use in transcriptome assembly has not been well demonstrated yet. Recently, the de novo assembly of human transcriptome has been reported using ABySS program.20 Here, we present a de novo assembly approach for transcriptome of a plant species using only short-read sequence data.
The present study has two major goals. First, we report a strategy for de novo assembly of transcriptome using short-read sequence data and effect of sequence quality and various parameters. Secondly, we report for the first time the complete transcriptome of chickpea, the legume crop plant. We have generated millions of sequence reads from chickpea transcriptome sequencing. A non-redundant set of transcripts have been generated and various analyses, including GC content analysis, sequence similarity/conservation with other plant species, functional categorization and identification of simple sequence repeats (SSRs) have been done. Our data provide a very useful genomic resource for future studies in chickpea.
2. Materials and methods
2.1. Plant material
Chickpea (C. arietinum L. genotype ICC4958) seeds procured from ICRISAT, Hyderabad, India, were grown as described.21 Root and shoot tissue samples were collected from the 15-day-old seedlings grown in autoclaved mixture (1:1) of agropeat and vermiculite in 3in. plastic pots at 22 ± 1°C in a culture room with a photoperiod of 14 h. The mature leaves and flower buds were collected from plants grown in the field. At least three independent biological replicates of each tissue sample were harvested and immediately frozen in liquid nitrogen.
2.2. RNA isolation and quality controls
Total RNA was extracted from all the tissue samples using TRI Reagent (Sigma Life Science, USA) according to manufacturer's instructions. The quality and quantity of each RNA sample was assessed as described previously.21 Only the RNA samples with 260 of 280 ratio from 1.9 to 2.1, 260 of 230 ratio from 2.0 to 2.5 and RIN (RNA integrity number) more than 8.0, were used for the analysis.
2.3. Illumina sequencing and quality controls
Three cDNA libraries were generated using mRNA-Seq assay for transcriptome sequencing on Illumina Genome Analyzer II platform. One paired-end (PE) cDNA library was generated from the pooled total RNA of shoot, root, mature leaf and flower buds in equal quantity and sequencing was done in one lane to generate 72 bp PE reads. Two cDNA libraries were generated one each from total RNA of root and shoot tissues and sequencing was done in one lane each to generate 51 bp single-end (SE) reads. The library construction and sequencing was performed by commercial service providers (PE, Genotypic Technology, Bangalore, India; SE, BC Cancer Agency Genome Sciences Centre, Vancouver, Canada). The sequence data generated in this study have been deposited at NCBI in the Short Read Archive database under the accession number SRA023503 (experiment accession numbers SRX025413 and SRX025414 for PE and SE read sequencing, respectively). Various quality controls, including filtering of high-quality reads based on the score value given in fastq files, removal of reads containing primer/adaptor sequences and trimming of read length were done using in-house tool kit (Patel and Jain, unpublished). We evaluated the effect of sequence quality on the de novo assembly of sequence reads.
2.4. De novo assembly
All the assemblies were performed on a server with 48cores and 128GB random access memory. We used various programs for de novo assembly of the PE and SE sequence reads to generate a non-redundant set of transcripts. Among the various programs available, we validated publicly available program, Velvet (version 0.7.62; http://www.ebi.ac.uk/~zerbino/velvet/), which have been developed for assembly of short reads using de Bruijn graph algorithm.17 Various assembly parameters were also optimized for best results. In addition, we used other publicly available programs, including Oases (version 0.1.8; http://www.ebi.ac.uk/~zerbino/oases/), ABySS (version 1.1.2; http://www.bcgsc.ca/platform/bioinfo/software/abyss)18 and SOAPdenovo (version 1.04; http://soap.genomics.org.cn/soapdenovo.html), and commercially available CLC Genomics workbench (version 3.7.1), which have also been developed for de novo assembly of short reads, to obtain best assembly results with our data set.
2.5. Similarity search and functional annotation
The proteome data sets for all the completely sequenced plant genomes so far were downloaded from their respective genome project websites. For generating non-redundant unigene data sets from various legume species, including Glycine max (soybean), Medicago truncatula, Lotus japonicus, Vigna unguiculata and Pisum sativum, all the available EST and mRNA sequences were downloaded from Genbank and assembled using TGI Clustering Tool (TGICL) after removing/trimming contaminating vector sequences and short reads (<100 bp) using SeqClean. The parameters used for assembly were more than 95% identity over a minimum of 40 bases with maximum of 20 bases of unmatched overhangs at sequence end. The chickpea transcripts were searched against proteome sequences and legume unigenes sets using BLASTX and TBLASTX searches, respectively, with an expect (E)-value cut-off of ≤1E−05 to reveal sequence conservation.
To deduce the putative function, chickpea non-redundant transcript data set was subjected to BLASTX analysis against the non-redundant protein database of UniProt and all annotated protein sequences of Arabidopsis (available at The Arabidopsis Information Resource). The results of only the best hit were extracted and the hits with an E-value ≤ 1E−05 were considered to be significant. The GOSlim terms for molecular function, biological process, and cellular component categories associated with the best BLASTX hit with Arabidopsis protein were assigned to the corresponding chickpea transcript. For the identification of transcription factor families represented in chickpea transcriptome, the chickpea transcripts were searched against all the transcription factor protein sequences at Plant transcription factor database (PlnTFDB; http://plntfdb.bio.uni-potsdam.de/v3.0/downloads.php) using BLASTX with an E-value cut-off of ≤1E−05.
2.6. GC content analysis and SSRs identification
GC content analysis was done using in-house perl scripts. The perl script program MISA (MIcroSAtellite; http://pgrc.ipk-gatersleben.de/misa/) was used for identification of SSRs. The repeats of mono-nucleotide more than 10 times, di-nucleotides repeats more than 6 times, tri-, tetra-, penta- and hexa-nucleotide repeats more than 5 times were considered as search criteria in MISA script.
2.7. Mapping of sequence reads onto chickpea transcripts
We mapped all the reads from three experiments onto the non-redundant set of transcripts to quantify the abundance of transcripts assembled, using CLC Genomics Workbench and Maq (v0.7.1; http://maq.sourceforge.net/index.shtml) softwares and the number of reads and reads per million (rpm) corresponding to each transcript were determined. In addition, the coverage of each transcript was determined in terms of number of reads per kilobase per million (rpkm).
3. Results
3.1. Sequencing of chickpea transcriptome
We generated a total of 134 954 354 sequence reads, including 65 900 072 PE sequence reads (32 950 036 from each end) each 72 bp in length and 69 054 282 SE sequence reads each 51 bp in length, encompassing about 21GB of sequence data in fastq format (Table 1). We filtered the sequence data for low-quality reads at high stringency (reads with more than 30% of bases with Phred quality score of ≤20) and reads containing primer/adaptor sequence. This resulted in a total of 106 660 317 (79%) high-quality sequence reads, including 50 523 492 (76.7%) PE sequence reads each 72 bp in length and 56 136 825 (81.3%) SE sequence reads each 51 bp in length (Table 1). After filtering, the average quality score increased significantly at each base position of the sequence reads (Supplementary Fig. S1). The average quality score was more than 30 at each base position in all the three data sets except for last seven bases at the 3′ end in PE data set. The final data set comprising ∼107million very high-quality reads was used for optimization of de novo assembly and analysis of chickpea transcriptome (Supplementary Fig. S2).
Table 1.
Summary of data generated for chickpea transcriptome
| Library | Total no. of reads | Fastq file size (GB) | No. of reads after filtering low-quality reads | No. of reads after removing primer/adapter containing reads |
|---|---|---|---|---|
| Pooled (72 bp PE) | 65 900 072 | 11.44 | 52 430 156 | 50 523 492 |
| Root (51 bp SE) | 31 028 774 | 4.06 | 24 671 859 | 24 670 440 |
| Shoot (51 bp SE) | 38 025 508 | 5.17 | 31 468 818 | 31 466 385 |
| Total | 134 954 354 | 20.67 | 108 570 833 | 106 660 317 |
3.2. De novo assembly
The de novo assembly of chickpea transcriptome was optimized after assessing the effect of various assembly parameters, trimming bases at sequence read ends and different assembly programs as described below.
3.2.1. Assessment of the effect of assembly parameters
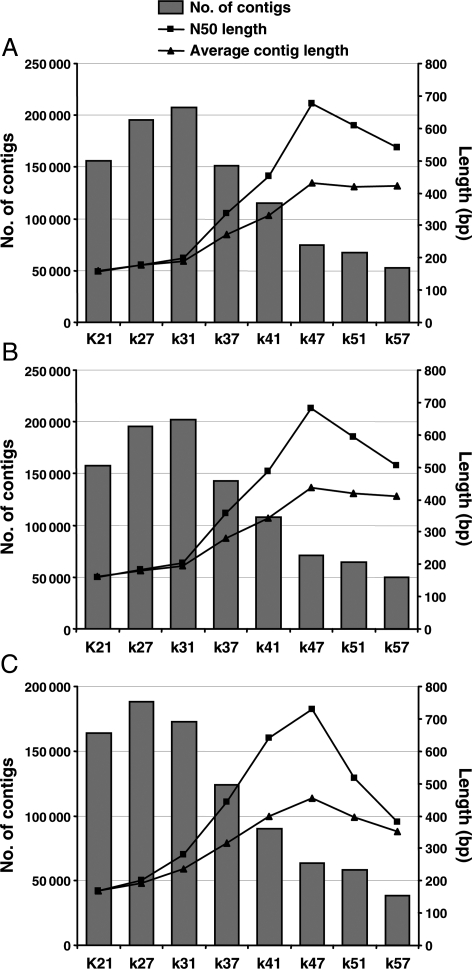
The untrimmed high-quality sequence reads were assembled using Velvet program at different k-mer length of 21, 27, 31, 37, 41, 47, 51 and 57. We analysed various output parameters like number of used reads, nodes, total number of contigs, contigs longer than 100 bp, N50 length, longest contig length and average contig length as a function of k-mer length (Supplementary Table S1A; Fig. 1). The results suggested that k-mer length affects inversely to the number of contigs. We found the best assembly to be that for k = 47, as it resulted in highest N50 length of 675 bp, largest contig length of 7827 bp and largest average contig length of 432 bp (Fig. 1A). The assembly resulted in a total of 74 651 contigs of at least 100 bp length. The total number of reads used for the assembly was also highest (73.6%) for k = 47 (Supplementary Table S1). We assessed the effect of other parameters such as insert length and expected coverage on Velvet assembly at different k-mer length and did not find any significant effect (data not shown).
Figure 1.
Comparison of de novo assembly of three data sets using Velvet program as a function of k-mer length. The three data sets include (A) untrimmed data set (72 bp PE and 51 bp SE reads), (B) trimmed data set 1 (70 bp PE and 50 bp SE reads) and (C) trimmed data set 2 (65 bp PE and 50 bp SE reads). The bars indicate number of contigs 100 bp or longer (left axis). The lines indicate N50 length (rectangles) and average contig length (triangles) in bp (right axis).
3.2.2. Assessment of the effect of trimming
To improve the quality of assembly and observe the effect of trimming of low-quality bases at the end of reads, we generated two trimmed data sets; first data set containing 70 bp PE (2 bp trimmed from 3′ end) and 50 bp SE (1 bp trimmed from 3′ end) sequence reads and second data set containing 65 bp PE (7 bp trimmed from 3′ end) and 50 bp SE (1 bp trimmed from 3′ end) sequence reads. We then assembled both these data sets also with different k-mer length using Velvet and noted various output parameters (Supplementary Table S1B and C; Fig. 1B and C). For first trimmed data set, the best assembly was for k = 47 with improved highest N50 length of 683 bp, largest contig length of 7666 bp and average contig length of 437 bp. The total number of contigs generated with at least 100 bp in length was 71 217 using 75% of the total reads. For the second trimmed data set, although the largest contig length was highest for k = 41 (10 616 bp), N50 length of 730 bp and average contig length of 453 bp were better for k = 47. The total number of contigs generated with at least 100 bp in length was 63 365 at k = 47 using highest number of total reads (77.8%). Taken together, we considered the assembly results of second trimmed data set for k = 47 as the best. The results show that the trimming of low-quality bases at the sequence read ends improved the assembly significantly.
3.2.3. Assessment of assembly programs
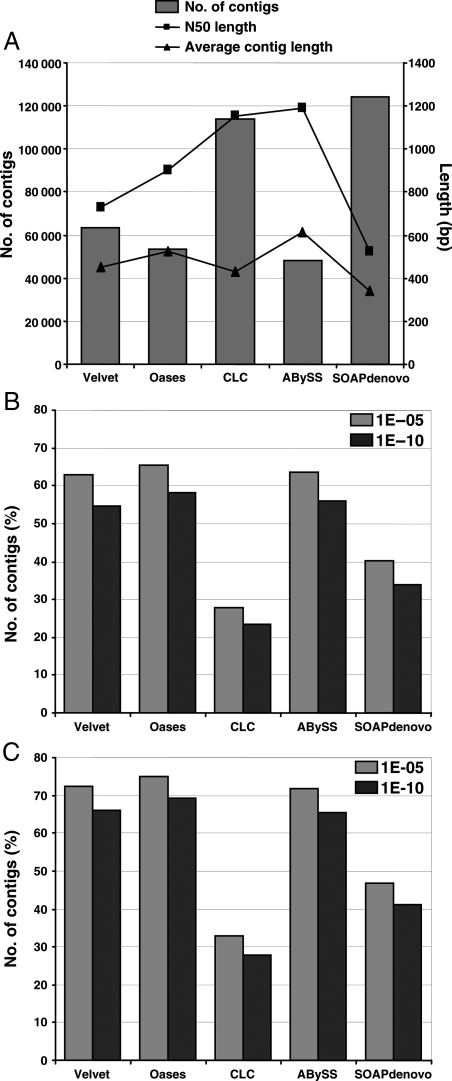
It has been suggested that assembly of Velvet followed by Oases yields better contigs/transcripts. The Oases program has been developed specifically for the de novo assembly of transcriptomes using short reads, which takes the assembly generated by Velvet as input and exploits the read sequence and pairing information to produce transcript isoforms. We performed assembly of contigs generated by Velvet for the second trimmed data set (k = 47) into transcripts using Oases with default parameters. This resulted in a total number of 59 178 transcript isoforms (≥100 bp in length). The number of reads used increased by 5% in Oases (82.8%) when compared with Velvet (77.8%). Among these, 4232 transcripts were represented by 10 001 transcript isoforms, which might represent alternative splicing events. From the 59 178 transcript isoforms obtained by Oases assembly, a set of 53 409 non-redundant transcripts (including only the largest transcript isoform) was obtained with N50 length of 900 bp, largest contig length of 8173 bp and average contig length of 523 bp (Fig. 2A). In addition, we also performed assembly of above three data sets (untrimmed data set, trimmed data set 1 and trimmed data set 2) using CLC Genomics workbench. We obtained the best assembly results with the second trimmed data set with a total number of 113 893 contigs (≥100 bp in length) with N50 length of 1151 bp, largest contig length of 15 684 bp and average contig length of 428 bp (Supplementary Table S2; Fig. 2A). Although N50 length was better in CLC Genomics Workbench, the average contig length was lower than that of Oases and total number of contigs generated was also much higher than expected (Fig. 2A). Further, the assembly of second trimmed data set using ABySS (k = 47) and SOAPdenovo (k = 31) programs generated 48 185 and 124 160 contigs, respectively, of at least 100 bp length. The N50 and average lengths for contigs generated by ABySS were 1192 and 613 bp, respectively, and that of contigs generated by SOAPdenovo were 525 and 340 bp, respectively (Fig. 2A).
Figure 2.
Comparison and validation of de novo assembly using various assembly programs. (A) Comparison of de novo assembly of trimmed data set 2 (65 bp PE and 50 bp SE reads) using Velvet, Oases, CLC Genomics Workbench, ABySS and SOAPdenovo. The bars indicate number of contigs 100 bp or longer (left axis). The lines indicate N50 length (rectangles) and average contig length (triangles) in bp (right axis). (B and C) Percentage of contigs generated using Velvet, Oases, CLC Genomics Workbench, ABySS and SOAPdenovo showing significant hits with Arabidopsis (B) and soybean (C) proteins at different E-value cut-offs.
Further, the validation of assembly output was done by BLASTX search of contigs/transcripts generated by various programs against Arabidopsis and soybean annotated proteomes. The significant hits were identified at different E-value cut-offs. The largest number of transcripts generated by Oases assembly showed significant similarity with Arabidopsis and soybean proteins at all E-value cut-offs when compared with the contigs generated by Velvet, CLC Genomics Workbench, ABySS and SOAPdenovo programs (Fig. 2B and C). Although the contigs generated by ABySS showed higher N50 and average lengths, lesser number and percentage of contigs showed significant similarity with Arabidopsis and soybean proteins. Overall, taken together, the assembly of second trimmed short-read data set obtained by Oases was found better than others and this non-redundant transcript data set (53 409) was analysed further. The statistics of the final assembly obtained from Oases program are given in Table 2. The comparative analysis of all (32 747) the ESTs and ESTs reported in various individual transcriptome studies,6–8 available at NCBI [after removing/trimming contaminating vector sequences and short reads (<100 bp) using SeqClean] with the above non-redundant transcript data set of chickpea showed that most (79% of the total and 74–88% of the individual EST data set reported in previous studies) of the ESTs are represented in our data set showing ≥90% identity over a length of ≥100 bp.
Table 2.
Statistics of non-redundant set of chickpea transcripts obtained from Oases assembly
| Total number of reads | 106 660 317 |
| Number of used reads | 88 337 267 |
| Number of unused reads | 18 323 050 |
| Number of transcript isoforms (≥100 bp) | 59 178 |
| Number of non-redundant transcripts (≥100 bp) | 53 409 |
| Total size of transcriptome (bp) | 27 950 383 |
| N50 length (bp) | 900 |
| Average contig length (bp) | 523 |
| Largest contig length (bp) | 8173 |
| Average coverage (rpkm) | 25.7 |
| Average number of reads per transcript | 1616.7 |
bp, base pair; rpkm, reads per kilo base per million.
3.3. GC content analysis of chickpea transcriptome
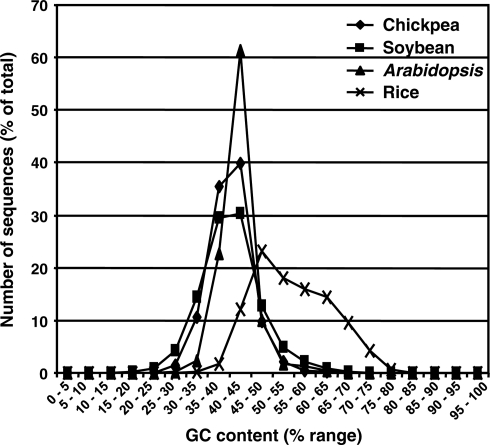
GC content (ratio of guanine and cytosine) of all the chickpea transcripts along with soybean (legume reference), Arabidopsis (dicot reference) and rice (monocot reference) was determined. The average GC content of chickpea transcripts (40.3%) and soybean unigenes (40.9%) was little lower than that of Arabidopsis (42.5%). The average GC content in rice was much higher (55%) as reported previously as well.22 Although the average GC contents of chickpea and Arabidopsis were comparable, chickpea has a higher proportion of transcripts with GC content in range of 35–40% but lower proportion of transcripts with high GC content in range of 40–45% (Fig. 3). In addition, the range of GC content was broader in chickpea and soybean when compared with Arabidopsis (Fig. 3). A similar observation was found for the unigene sets from other legume species as well (data not shown).
Figure 3.
GC content analysis of chickpea transcripts. The average GC content of each transcript for chickpea, soybean, Arabidopsis and rice was calculated and percentage of transcripts with GC content within a range are represented.
3.4. Identification of SSRs
The transcript/EST-based markers are important resource for determining functional genetic variation.23 Among the various molecular markers, SSRs are highly polymorphic, easier to develop and serve as rich resource of diversity. For identification of SSRs, all the chickpea transcripts were searched with perl script MISA. We identified a total of 4816 SSRs in 4180 (7.8%) transcripts of chickpea with frequency of one SSR per 5.80 kb of the sequence (Table 3). The mono-nucleotide SSRs represented the largest fraction (41.9%) of SSRs identified followed by tri-nucleotide (36.1%) and di-nucleotide (19.3%) SSRs. Although only a small fraction of tetra- (50), penta- (22) and hexa-nucleotide (58) SSRs were identified in chickpea transcripts, the number is quite significant.
Table 3.
Statistics of SSRs identified in chickpea transcripts
| SSR mining | |
| Total number of sequences examined | 53 409 |
| Total size of examined sequences (bp) | 27 950 383 |
| Total number of identified SSRs | 4816 |
| Number of SSR containing sequences | 4180 (7.8%) |
| Number of sequences containing more than one SSR | 534 |
| Number of SSRs present in compound formation | 282 |
| Frequency of SSRs | One per 5.80 kb |
| Distribution of SSRs in different repeat types | |
| Mono-nucleotide | 2020 (41.9%) |
| Di-nucleotide | 930 (19.3%) |
| Tri-nucleotide | 1736 (36.1%) |
| Tetra-nucleotide | 50 (0.010%) |
| Penta-nucleotide | 22 (0.005%) |
| Hexa-nucleotide | 58 (0.012%) |
3.5. Sequence similarity of chickpea transcripts with other plants
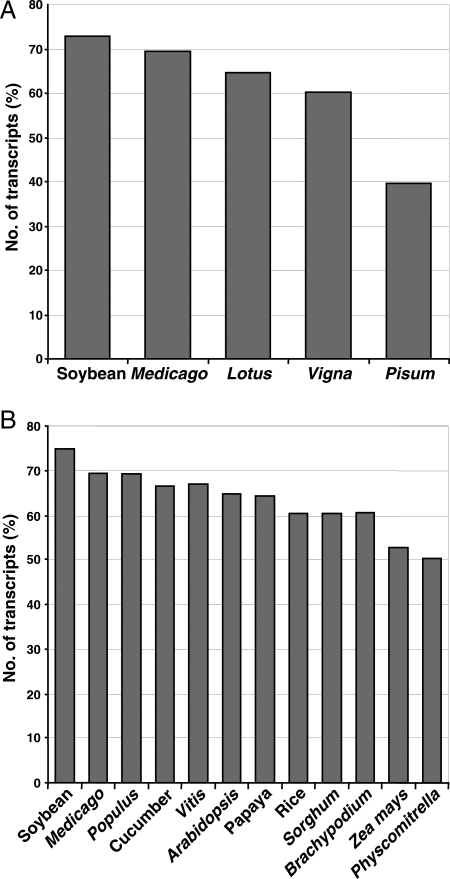
The transcript set of chickpea was analysed for similarity/sequence conservation against the unigene data sets of various legumes species namely soybean, Medicago, Lotus, Vigna and Pisum using TBLASTX search. An E-value cut-off threshold of ≤1E−05 was considered to define a significant hit. The largest number (72.4%) of chickpea transcripts showed significant similarity with soybean unigenes followed by Medicago (69.5%), Lotus (65.5%), Vigna (60.1%) and the least similarity with Pisum (39.3%; Fig. 4A). Overall, a total of 43 516 (81.5%) of the chickpea transcripts showed significant similarity with at least one of the other legume unigenes. Likewise, we analysed the sequence conservation of chickpea transcripts with proteomes of all sequenced plant species. A total of 42 012 (79%) transcripts exhibited significant similarity with at least one of the predicted protein from sequenced plants. The largest number (75%) of chickpea transcripts showed significant similarity with soybean followed by Medicago (69%) and least with Physcomitrella (50%; Fig. 4B). As expected, lesser number of chickpea transcripts showed significant similarity with monocots (52–61%) when compared with dicots (65–75%). Although a large number of the chickpea transcripts showed significant similarity with predicted proteins from legumes, the extent of coverage of the coding region was quite less than expected. Only 28.4 and 29.5% of the transcripts which showed significant similarity covered ≥50% of the coding region of the predicted proteins from soybean and Medicago, respectively. Considering a high degree of conservation among legumes, it may be assumed that the assembly of chickpea transcriptome may further be improved as more sequence data become available.
Figure 4.
Sequence conservation of chickpea transcripts with other plant species. (A) Sequence conservation of chickpea transcripts with putative transcript consensus (TC) sequences of various legume species. (B) Sequence conservation of chickpea transcripts with annotated proteins of completely sequenced plant species. The percentage of transcripts showing significant similarity (E-value ≤ 1E−05) in TBLASTX (A) and BLASTX (B) searches are shown.
Further, the combined analysis of BLAST results with legume unigenes and plant proteomes revealed that a total of 45 636 (85.5%) transcripts were conserved in chickpea showing significant similarity with at least one sequence. Among these, 3624 (6.8%) transcripts were conserved only in legumes representing legume-specific genes. However, 7773 (14.5%) transcripts did not show significant similarity with any of the data set analysed, and may represent chickpea-specific genes.
3.6. Functional annotation and characterization of chickpea transcripts
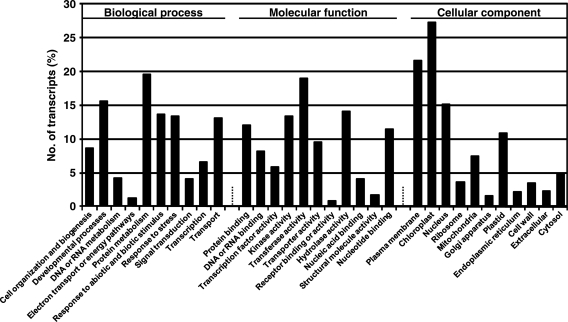
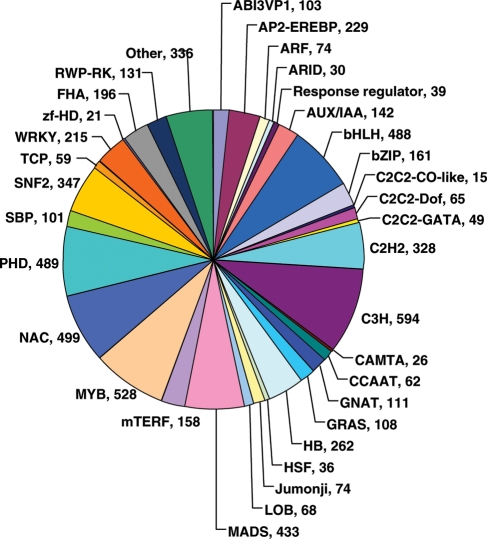
To identify the putative function of chickpea transcripts, they were compared against the non-redundant protein sequences available at UniProt database and Arabidopsis proteins using BLASTX search. A total of 23 864 (44.7%) and 34 993 (65.5%) chickpea transcripts showed significant hit with UniProt and Arabidopsis proteins, respectively. Together, 35 279 (66.1%) transcripts showed significant hit with at least one UniProt or Arabidopsis protein. Broadly, the putative orthologs of genes involved in various pathways and cellular processes were found to be conserved in chickpea. In addition, many chickpea transcripts showed homology to uncharacterized proteins annotated as unknown, hypothetical and expressed proteins as well. Further, GO terms were assigned to chickpea transcripts, which showed significant similarity with Arabidopsis proteins annotated with GO terms. A total of 34 676 (64.9%) transcripts were assigned at least one GO term, among which 31 250 were assigned at least one GO term in biological process category, 31 598 in molecular function category and 30 264 in cellular component category. Among the various biological processes, ignoring unknown and other biological process categories, protein metabolism (19.5%) and developmental processes (15.6%) were most highly represented (Fig. 5). The genes involved in other important biological processes such as response to abiotic and biotic stimulus/stress, transport, transcription and signal transduction, were also identified through GO annotations. Similarly, transferase activity and hydrolase activity were most represented among the various molecular functions, and chloroplast and plasma membrane were most represented among the cellular component categories (Fig. 5). Further, we identified transcription factor encoding transcripts by sequence comparison to known transcription factor gene families. In total, 6577 putative chickpea transcription factor genes, distributed in at least 57 families, were identified representing 12.3% of chickpea transcripts (Fig. 6). The overall distribution of transcription factor encoding transcripts among the various known protein families is very similar with that of soybean and other legumes as predicted earlier.24,25 However, a few families showed the events of expansion (for example, Aux/IAA-ARF, bHLH, C3H, MADS, NAC, PHD and RWP-RK etc.) and contraction (for example, C2C2 zinc finger, CCAAT, LIM and MYB etc.) indicating their evolutionary significance (Supplementary Table S3). In fact, the number of predicted transcription factor encoding genes in Medicago and Lotus are very less when compared with soybean.24,25 As the complete genome sequence is available only for soybean as of now, the complete picture about evolution of various transcription factor families will emerge once the complete genome sequences and analyses thereof will be available for other legume species too.
Figure 5.
Functional annotation of chickpea transcripts. GOSlim term assignment to the chickpea transcripts in different categories of biological process, molecular function and cellular component.
Figure 6.
Distribution of chickpea transcripts in different transcription factor families.
3.7. Quantification of chickpea transcripts
The digital expression profiling, also called RNA-Seq, is a powerful and efficient approach for gene expression analysis.26,27 The mapping of all the reads onto the non-redundant set of chickpea transcripts revealed that the number of reads corresponding to each transcript ranged from 14 (0.16 rpm) to 270 894 (3,137.3 rpm) with an average of 1617 reads (18.7 rpm) per transcript, indicating a very wide range of expression levels of chickpea transcripts (Supplementary Table S4). It also indicates that very low expressed chickpea transcripts are also represented in our assembly. The minimum coverage (rpkm) of a chickpea transcript was 1.2 and maximum of 9015.1 with an average of 25.7 (Table 2). Further, we analysed the expression of chickpea transcripts in root, shoot and pooled (root, shoot, mature leaf and flower bud) tissue samples. The mapping of reads on the chickpea transcript data set revealed that a total of 1974 and 1174 transcripts are not represented in the root and shoot tissue sequence data sets, respectively. Among these, 420 transcripts have no read mapped from root and shoot data sets, indicating their expression in mature leaf and/or flower buds (Supplementary Table S4). Further, another 250 transcripts were found to have root-specific expression, as they have no read mapped from shoot data set and at least 3 rpm mapped from root data set (Supplementary Table S4). Likewise, another 217 transcripts were found to have shoot-specific expression, as they have no read mapped from root data set and at least 3 rpm mapped from shoot data set (Supplementary Table S4). However, their expression in other chickpea tissues not analysed in this study is not ruled out.
4. Discussion
The transcriptome sequencing enables various functional genomic studies for an organism. Although several high throughput technologies have been developed for rapid sequencing and characterization of transcriptomes, expressed sequence data are still not available for many organisms, including crop plants. The next generation sequencing technologies provide a low cost, labour saving and rapid means of transcriptome sequencing and characterization.14 Similar to sequencing technologies, many bioinformatics tools have also been developed for the short-read sequence data assembly and analysis,17,18 but the requisite knowledge is very limited. The de novo assembly of short reads without a known reference is considered difficult.28 Therefore, the use of more expensive 454 Life Sciences (Roche) technology is used for non-model organisms, which produces longer sequence reads.15 However, the de novo assembly of transcriptomes using short reads has also received attention.20,29 In this study, we demonstrate a strategy for de novo assembly of transcriptome using short reads for a non-model crop plant, chickpea, for which sequence data is very limited so far in the public databases. We showed that there is significant effect of the assembly program parameters and sequence quality on the assembly output. A larger N50 length and average length are considered indicative of better assembly. Our results show that N50 length of the contigs generated using Velvet assembly program varied greatly as a function of k-mer length and increased from 675 bp for untrimmed data set to 730 bp after trimming low-quality bases. Hence, we suggest that the optimization of program parameters and trimming of low-quality bases at the ends of sequence reads might improve the assembly output significantly. In addition, the validation of different assembly programs is also required to get the optimum results. We found the assembly of Velvet followed by Oases program better than that of Velvet alone, CLC Genomics Workbench, ABySS and SOAPdenovo programs based on various assessment parameters such as N50 length, average contig length and sequence similarity with closely related species.
Chickpea is one of the most important legume crop plants rich in proteins, carbohydrates and other nutrients, which makes it very important target for genomic studies.1,2 For the model legumes such as soybean, Medicago and Lotus, genome sequencing has been nearly completed and a vast collection of ESTs are available for functional genomic studies.25,30–32 However, very few genomic resources, including genome sequence, EST sequences and molecular markers are available for chickpea so far when compared with other legumes. We have generated more than 100million sequence reads for chickpea and report a non-redundant set of 53 409 transcripts representing about 28 Mb sequence and 3.8% of the chickpea genome. The coverage of chickpea transcripts was quite high (average of 25.7 rpkm), which is very crucial for quality and length of transcripts obtained. The GC content analysis revealed that chickpea transcripts have a low GC content similar to other dicots. However, the GC content range was broader when compared with other dicots which cover a narrow GC range.33 The GC content analysis provide insights into various aspects related to genome of an organism, including evolution, gene structure (intron size and number), thermostability and gene regulation.22,34,35 We identified a large number of SSRs in the chickpea transcripts. The number of tri-nucleotide SSRs was much higher than di-nucleotide SSRs. Earlier studies have also reported the higher number of tri-nucleotide SSRs when compared with di-nucleotide SSRs in ESTs than genomic sequences.23,36 However, recently the larger number of di-nucleotide SSRs than tri-nucleotide SSRs has been reported in pigeonpea ESTs.37 The frequency and distribution of SSRs have been proposed to be dependent on various factors such as size of data set, tools and criteria used.23 The identification of SSRs provides a very cost-effective option to develop functional markers for various marker-assisted breeding purposes.
The analysis of sequence conservation helps in transfer of knowledge from model plants to chickpea for functional genomic studies. A large number (60–72%) of chickpea transcripts showed significant similarity with legumes at protein level as expected except for Pisum, indicating that their function might also be conserved. The low similarity with Pisum unigenes may be due to the availability of lesser number of sequences. The low similarity of chickpea transcripts with monocot proteomes when compared with dicot proteomes is also not unexpected due to the phylogenetic divergence of monocots and dicots during evolution. A significant number of transcripts were found to be conserved only in legumes, which may perform legume-specific functions. Interestingly, about 15% of the transcripts did not show significant homology with any other sequences, which may be novel and perform species-specific functions. The lineage- and species-specific genes have been identified in other plant species, including legumes as well.38–40 The study of these genes will be very important to dissect the lineage- or species-specific cellular processes and study evolutionary processes such as speciation and adaptation. Further, the chickpea transcripts were found to belong to various functional categories conserved in other plants also. However, overrepresentation of few functional categories might provide a clue towards specific functions/pathways operative in legumes or chickpea. Likewise, the conservation of transcription factor families indicates the presence of conserved gene regulatory machinery in chickpea. However, the lineage- and/or species-specific evolutionary expansion and contraction along with unique gene expression patterns of some of transcription factors may contribute to the legume-specific traits.
In conclusion, we have demonstrated the use of short-read sequence data to rapidly characterize a draft transcriptome of an organism. The strategy of de novo assembly described here can be potentially used for any species. In addition, our study contributes a significant non-redundant set of 53 409 transcripts in chickpea. The detailed analyses of the data set has provided several important features of chickpea transcriptome such as GC content, conserved genes across legumes and other plant species, assignment of functional categories and identification of SSRs. It is anticipated that this study is a significant contribution towards development of genomic resources for chickpea and will accelerate functional genomic studies and breeding programmes.
Supplementary Data
Supplementary Data are available at www.dnaresearch.oxfordjournals.org.
Funding
This study was supported financially by the Department of Biotechnology, Government of India, New Delhi, under the Next Generation Challenge Programme on Chickpea Genomics.
References
- 1.Graham P.H., Vance C.P. Legumes: importance and constraints to greater use. Plant Physiol. 2003;131:872–7. doi: 10.1104/pp.017004. doi:10.1104/pp.017004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Millan T., Clarke H.J., Siddique K.H.M., et al. Chickpea molecular breeding: new tools and concepts. Euphytica. 2006;147:81–103. doi:10.1007/s10681-006-4261-4. [Google Scholar]
- 3.Varshney R.K., Graner A., Sorrells M.E. Genomics-assisted breeding for crop improvement. Trends Plant Sci. 2005;10:621–30. doi: 10.1016/j.tplants.2005.10.004. doi:10.1016/j.tplants.2005.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Varshney R.K., Hoisington D.A., Tyagi A.K. Advances in cereal genomics and applications in crop breeding. Trends Biotechnol. 2006;24:490–9. doi: 10.1016/j.tibtech.2006.08.006. doi:10.1016/j.tibtech.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Buhariwalla H.K., Jayashree B., Eshwar K., Crouch J.H. Development of ESTs from chickpea roots and their use in diversity analysis of the Cicer genus. BMC Plant Biol. 2005;5:16. doi: 10.1186/1471-2229-5-16. doi:10.1186/1471-2229-5-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao W.R., Wang X.S., Liu Q.Y., et al. Comparative analysis of ESTs in response to drought stress in chickpea (C. arietinum L.) Biochem. Biophys. Res. Commun. 2008;376:578–83. doi: 10.1016/j.bbrc.2008.09.030. doi:10.1016/j.bbrc.2008.09.030. [DOI] [PubMed] [Google Scholar]
- 7.Ashraf N., Ghai D., Barman P., et al. Comparative analyses of genotype dependent expressed sequence tags and stress-responsive transcriptome of chickpea wilt illustrate predicted and unexpected genes and novel regulators of plant immunity. BMC Genomics. 2009;10:415. doi: 10.1186/1471-2164-10-415. doi:10.1186/1471-2164-10-415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Varshney R.K., Hiremath P.J., Lekha P., et al. A comprehensive resource of drought- and salinity-responsive ESTs for gene discovery and marker development in chickpea (Cicer arietinum L.) BMC Genomics. 2009;10:523. doi: 10.1186/1471-2164-10-523. doi:10.1186/1471-2164-10-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jain D., Chattopadhyay D. Analysis of gene expression in response to water deficit of chickpea (Cicer arietinum L.) varieties differing in drought tolerance. BMC Plant Biol. 2010;10:24. doi: 10.1186/1471-2229-10-24. doi:10.1186/1471-2229-10-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mantri N.L., Ford R., Coram T.E., Pang E.C. Transcriptional profiling of chickpea genes differentially regulated in response to high-salinity, cold and drought. BMC Genomics. 2007;8:303. doi: 10.1186/1471-2164-8-303. doi:10.1186/1471-2164-8-303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Molina C., Rotter B., Horres R., et al. SuperSAGE: the drought stress-responsive transcriptome of chickpea roots. BMC Genomics. 2008;9:553. doi: 10.1186/1471-2164-9-553. doi:10.1186/1471-2164-9-553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shukla R.K., Raha S., Tripathi V., Chattopadhyay D. Expression of CAP2, an APETALA2-family transcription factor from chickpea, enhances growth and tolerance to dehydration and salt stress in transgenic tobacco. Plant Physiol. 2006;142:113–23. doi: 10.1104/pp.106.081752. doi:10.1104/pp.106.081752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tripathi V., Parasuraman B., Laxmi A., Chattopadhyay D. CIPK6, a CBL-interacting protein kinase is required for development and salt tolerance in plants. Plant J. 2009;58:778–90. doi: 10.1111/j.1365-313X.2009.03812.x. doi:10.1111/j.1365-313X.2009.03812.x. [DOI] [PubMed] [Google Scholar]
- 14.Morozova O., Hirst M., Marra M.A. Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genomics Hum. Genet. 2009;10:135–51. doi: 10.1146/annurev-genom-082908-145957. doi:10.1146/annurev-genom-082908-145957. [DOI] [PubMed] [Google Scholar]
- 15.Vera J.C., Wheat C.W., Fescemyer H.W., et al. Rapid transcriptome characterization for a nonmodel organism using 454 pyrosequencing. Mol. Ecol. 2008;17:1636–47. doi: 10.1111/j.1365-294X.2008.03666.x. doi:10.1111/j.1365-294X.2008.03666.x. [DOI] [PubMed] [Google Scholar]
- 16.Zeng S., Xiao G., Guo J., et al. Development of a EST dataset and characterization of EST-SSRs in a traditional Chinese medicinal plant, Epimedium sagittatum (Sieb. Et Zucc.) Maxim. BMC Genomics. 2010;11:94. doi: 10.1186/1471-2164-11-94. doi:10.1186/1471-2164-11-94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–9. doi: 10.1101/gr.074492.107. doi:10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simpson J.T., Wong K., Jackman S.D., Schein J.E., Jones S.J., Birol I. ABySS: a parallel assembler for short read sequence data. Genome Res. 2009;19:1117–23. doi: 10.1101/gr.089532.108. doi:10.1101/gr.089532.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jackson B.G., Schnable P.S., Aluru S. Parallel short sequence assembly of transcriptomes. BMC Bioinformatics. 2009;10(Suppl 1):S14. doi: 10.1186/1471-2105-10-S1-S14. doi:10.1186/1471-2105-10-S1-S14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Birol I., Jackman S.D., Nielsen C.B., et al. De novo transcriptome assembly with ABySS. Bioinformatics. 2009;25:2872–7. doi: 10.1093/bioinformatics/btp367. doi:10.1093/bioinformatics/btp367. [DOI] [PubMed] [Google Scholar]
- 21.Garg R., Sahoo A., Tyagi A.K., Jain M. Validation of internal control genes for quantitative gene expression studies in chickpea (Cicer arietinum L.) Biochem. Biophys. Res. Commun. 2010;396:283–8. doi: 10.1016/j.bbrc.2010.04.079. doi:10.1016/j.bbrc.2010.04.079. [DOI] [PubMed] [Google Scholar]
- 22.Carels N., Bernardi G. Two classes of genes in plants. Genetics. 2000;154:1819–25. doi: 10.1093/genetics/154.4.1819. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Varshney R.K., Graner A., Sorrells M.E. Genic microsatellite markers in plants: features and applications. Trends Biotechnol. 2005;23:48–5. doi: 10.1016/j.tibtech.2004.11.005. doi:10.1016/j.tibtech.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 24.Libault M., Joshi T., Benedito V.A., Xu D., Udvardi M.K., Stacy G. Legume transcription factor genes: what makes legumes so special. Plant Physiol. 2009;151:991–1001. doi: 10.1104/pp.109.144105. doi:10.1104/pp.109.144105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schmutz J., Cannon S.B., Schlueter J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. doi:10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 26.Velculescu V.E., Kinzler K.W. Gene expression analysis goes digital. Nat. Biotechnol. 2007;25:878–80. doi: 10.1038/nbt0807-878. doi:10.1038/nbt0807-878. [DOI] [PubMed] [Google Scholar]
- 27.Wang Z., Gerstein M., Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat. Rev. Genet. 2009;10:57–63. doi: 10.1038/nrg2484. doi:10.1038/nrg2484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schuster S.C. Next-generation sequencing transforms today's biology. Nat. Methods. 2008;5:16–8. doi: 10.1038/nmeth1156. doi:10.1038/nmeth1156. [DOI] [PubMed] [Google Scholar]
- 29.Gibbons J.G., Janson E.M., Hittinger C.T., Johnston M., Abbot P., Rokas A. Benchmarking next-generation transcriptome sequencing for functional and evolutionary genomics. Mol. Biol. Evol. 2009;26:2731–44. doi: 10.1093/molbev/msp188. doi:10.1093/molbev/msp188. [DOI] [PubMed] [Google Scholar]
- 30.Cheung F., Haas B.J., Goldberg S.M., May G.D., Xiao Y., Town C.D. Sequencing Medicago truncatula expressed sequenced tags using 454 Life Sciences technology. BMC Genomics. 2006;7:272. doi: 10.1186/1471-2164-7-272. doi:10.1186/1471-2164-7-272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sato S., Nakamura Y., Kaneko T., et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–39. doi: 10.1093/dnares/dsn008. doi:10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cannon S.B., May G.D., Jackson S.A. Three sequenced legume genomes and many crop species: rich opportunities for translational genomics. Plant Physiol. 2009;151:970–7. doi: 10.1104/pp.109.144659. doi:10.1104/pp.109.144659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Carels N., Hatey P., Jabbari K., Bernardi G. Compositional properties of homologous coding sequences from plants. J. Mol. Evol. 1998;46:45–53. doi: 10.1007/pl00006282. doi:10.1007/PL00006282. [DOI] [PubMed] [Google Scholar]
- 34.Vinogradov A.E. DNA helix: the importance of being GC-rich. Nucleic Acids Res. 2003;31:1838–44. doi: 10.1093/nar/gkg296. doi:10.1093/nar/gkg296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhang L., Kasif S., Cantor C.R., Broude N.E. GC/AT-content spikes as genomic punctuation marks. Proc. Natl. Acad. Sci. USA. 2004;101:16855–60. doi: 10.1073/pnas.0407821101. doi:10.1073/pnas.0407821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Luo M., Dang P., Guo B.Z., et al. Generation of expressed sequence tags (ESTs) for gene discovery and marker development in cultivated peanut. Crop Sci. 2005;45:346–53. doi:10.2135/cropsci2005.0346. [Google Scholar]
- 37.Raju N.L., Gnanesh B.N., Lekha P., et al. The first set of EST resource for gene discovery and marker development in pigeonpea (Cajanus cajan L.) BMC Plant Biol. 2010;10:45. doi: 10.1186/1471-2229-10-45. doi:10.1186/1471-2229-10-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Graham M.A., Silverstein K.A., Cannon S.B., VandenBosch K.A. Computational identification and characterization of novel genes from legumes. Plant Physiol. 2004;135:1179–97. doi: 10.1104/pp.104.037531. doi:10.1104/pp.104.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Campbell M.A., Zhu W., Jiang N., et al. Identification and characterization of lineage-specific genes within the Poaceae. Plant Physiol. 2007;145:1311–22. doi: 10.1104/pp.107.104513. doi:10.1104/pp.107.104513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lin H., Moghe G., Ouyang S., et al. Comparative analyses reveal distinct sets of lineage-specific genes within Arabidopsis thaliana. BMC Evol. Biol. 2010;10:41. doi: 10.1186/1471-2148-10-41. doi:10.1186/1471-2148-10-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.