Abstract
Screening large numbers of target regions in multiple DNA samples for sequence variation is an important application of next-generation sequencing but an efficient method to enrich the samples in parallel has yet to be reported. We describe an advanced method that combines DNA samples using indexes or barcodes prior to target enrichment to facilitate this type of experiment. Sequencing libraries for multiple individual DNA samples, each incorporating a unique 6-bp index, are combined in equal quantities, enriched using a single in-solution target enrichment assay and sequenced in a single reaction. Sequence reads are parsed based on the index, allowing sequence analysis of individual samples. We show that the use of indexed samples does not impact on the efficiency of the enrichment reaction. For three- and nine-indexed HapMap DNA samples, the method was found to be highly accurate for SNP identification. Even with sequence coverage as low as 8x, 99% of sequence SNP calls were concordant with known genotypes. Within a single experiment, this method can sequence the exonic regions of hundreds of genes in tens of samples for sequence and structural variation using as little as 1 μg of input DNA per sample.
Keywords: next-generation sequencing, enrichment, capture, SNP, index
1. Introduction
Next-generation sequencing technology has the potential to allow sequencing of whole genomes to be carried out in standard molecular genetics laboratories. However, an important current application is the sequencing of specific genomic regions, e.g. genes with known or suspected mutations in patient samples. In order to sequence parts of the genome of interest, a number of target enrichment procedures have been developed. These include standard PCR, long-range PCR, nested patch PCR, template circularization, the use of gapped molecular inversion probes, microarray capture, in-solution capture and microdroplet-based PCR enrichment.1–9 All these methods work but each has its own advantages and disadvantages. A recent review on target enrichment strategies for next-generation sequencing, which discusses the methods listed above, concluded that the ability to combine DNA samples prior to enrichment would be an important advancement for targeted next-generation sequencing: ‘The logical extension of sample pooling is to perform multiplexed target enrichments in which many samples are barcoded before capture’.10 Here, we describe a method that delivers this advancement by combining DNA samples using indexes or barcodes prior to target enrichment.
We have adapted the indexing protocol published by Craig et al.11 and combined it with the Agilent Technologies SureSelect Target Enrichment System to develop a cost-efficient method for targeting smaller regions of the genome (e.g. 100 kb–1 Mb) in multiple DNA samples. SureSelect is a capture protocol based on the in-solution method developed by Gnirke et al.12 This method allows targeting of regions of the genome in custom designed reactions by using cRNA baits. By enabling simultaneous enrichment of multiple samples with no impact on individual sample identification for downstream analysis, this method significantly reduces the cost and time required for this type of experiment. Our method is highly suited to target enrichment of smaller genomic regions. The method can accurately detect SNPs at enriched target sites. Within a single sequencing experiment, it has the capacity to analyze the exonic regions of hundreds of genes in tens of samples for sequence variation using as little as 1 μg of input DNA.
2. Materials and methods
2.1. Enrichment reaction eArray design
We focused on two genes on chromosome 1 (PTBP2 and CDC42). The plan was to sequence the genes in full and include upstream and downstream regions. The target region for CDC42 was 108 139 bp, and for PTBP2, it was 184 850 bp. Bait libraries were designed and assessed for coverage across the target genomic regions using the Agilent eArray website (https://earray.chem.agilent.com/earray/). The online design recommends repeat masking of the target sequence in order to minimize off-target capture. With the default repeat masking option turned on, it was only possible to design target capture baits for just under 40% of CDC42 and just under 43% for PTBP2 (see cRNA baits RM track in Supplementary Fig. SA, e.g. of coverage across PTBP2 with default options). In order to try and increase the percentage of target capture, we adopted an alternative repeat masking protocol: the repeat masking constraints were relaxed to allow relatively unique baits to remain in the pool of target capture baits. This was achieved by designing baits using the Agilent eArray website to the target region with the repeat masking option turned off. The resulting baits were searched against the human genome reference (hg18) for similarity. If a bait mapped to more than one location with greater than 90% sequence identity using BLAST (∼12 mismatches across the bait), this bait was removed from the design. Using this protocol, we were able to design baits for just over 54% of CDC42 and 77% of PTBP2 (see cRNA baits SRM in Supplementary Fig. SA, e.g. of coverage across PTBP2 with our alternative relaxed repeat masking). Because the bait library was a fraction of the total possible for eArray design, we replicated it four times using 92.18% of the available space on the array.
2.2. Index design
The indexing methodology was taken from the study by Craig et al.11 The specific 6 bp indexes used in this study (listed in Supplementary Table SA) were from a group of 19 indexes that performed most reliably in that study (Fig. 2 in Craig et al.11). Each index is designed such that it can tolerate at least 1 bp change as a result of a sequencing error without mutating into one of the other indexes used in the study. This helps guard against potential mix-up of individual sample data.
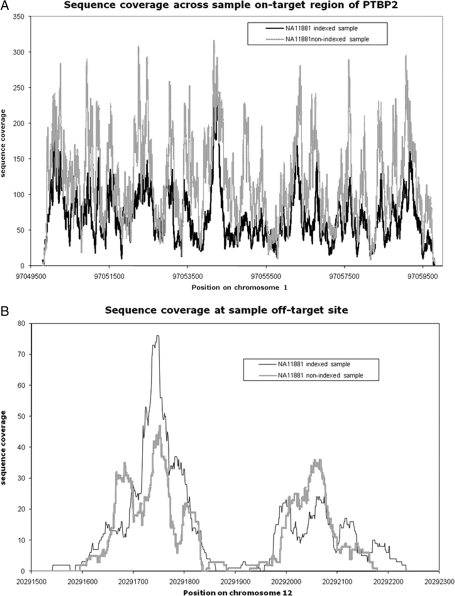
Figure 2.
Sequence coverage across on-target and off-target regions. Sequence coverage is plotted for the single non-indexed and indexed samples at an on-target site (PTBP2 on chromosome 1; A) and at an off-target site (chromosome 12; B). Inclusion of the index does not dramatically change the pattern of sequence coverage at on-target or off-target regions. The higher sequence coverage observed for the non-indexed sample compared with the indexed sample reflects the larger number of clusters that passed QC filtering during the sequence run (Supplementary Table SB).
2.3. Library preparation, target enrichment and sequencing of genomic DNA samples
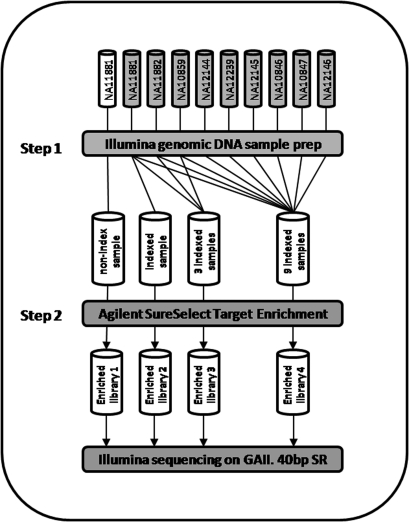
DNA from each of nine CEU HapMap samples (three trios) was used in this study. A total of four enriched sequencing libraries were generated using these samples (Fig. 1). Library 1 contained one target enriched non-indexed DNA sample. Library 2 contained one target enriched indexed DNA sample. Library 3 was a multiplex sequencing library of three equimolar indexed samples, which was enriched using one target enrichment reaction. Library 4 was a multiplex sequencing library of nine equimolar indexed samples, which was enriched using one target enrichment reaction. Figure 1 identifies the HapMap samples used in each lane of sequencing. The issue of potential differences in quantity of sequence reads between indexed samples due to the efficiency of indexed adapter ligation to input DNA, as previously reported by Craig et al.,11 was addressed in this study by multiplexing the indexed in equal quantities pre-enrichment.
Figure 1.
Experimental Design. Genomic DNA from nine HapMap samples was chosen for the study (three trio families). DNA from one of the samples (NA11881) was prepared twice (with and without an indexed adapter), target enriched and sequenced separately as single samples (non-indexed sample and one indexed sample in Step 1 and enriched libraries 1 and 2 in Step 2). One trio family (NA11881, NA11882 and NA10859; all indexed) was pooled after the Illumina genomic DNA sample prep and enriched together using one SureSelect enrichment reaction to produce the enriched library 3 sample. Indexed DNA from all nine samples was also pooled after the Illumina genomic DNA sample prep and enriched together using one SureSelect enrichment reaction to produce the enriched library 4 sample. Note: enriched libraries 3 and 4 were also sequenced using 80 bp reads to generate additional data for validation of the method for SNP detection.
The preparation of each sample is a two-step process; in the first step, the DNA is prepared as an Illumina sequencing library, and in the second step, the sequencing library is enriched for the desired target using the Agilent SureSelect enrichment protocol. The library preparation and enrichment methods were followed according to the Agilent Illumina Single-End Sequencing Platform Library Prep protocol (v1.2 April 2009) with the following modifications: (1) instead of shearing the DNA with a covaris system, a biorupter (Diagenode) was used. The samples were sonicated on high for 30 s and off for 30 s for a total 30 min with addition of ice after every 10 min to keep the samples cool. (2) For the indexed samples (libraries 2–4), the Illumina adapters were replaced with custom made indexed adapters supplied by IDT DNA. During the SureSelect enrichment process, blocking oligos are used (provided by Agilent) to temporarily block the Illumina adapter sequence and prevent off-target pull-down of genomic DNA due to similarity of sequence in the cRNA baits and Illumina adapter sequence. (3) For the pre-capture enrichment PCR, 1.3 µl of DNA was used as input with 11 cycles of PCR instead of 1 µl of input DNA and 14 cycles of PCR. (4) Post-enriched library DNA from the individual samples were combined (one, three or nine samples) to a total quantity of 500 ng per pool. This solution was allowed to evaporate off overnight instead of using a vacuum concentrator and resuspended in 3.4 µl of elution buffer to give a final concentration of ∼147 ng/µl. The individual sample combinations (one, three or nine-samples) were each enriched using one custom SureSelect enrichment reaction (library design ELID: 0236181). Target enriched libraries were stored at a stock concentration of 10 nM ready for sequencing. A total of 6/8 pM of target enriched libraries were sequenced on the Illumina Genome Analyzer II using 40 bp reads following the manufacturers protocol. Libraries 3 and 4 (three and nine samples) were also sequenced using 80 bp reads to generate additional data for the study.
2.4. Data analysis
The base sequence data were called from the image files with the Illumina Bustard.py script and the Illumina GA pipeline version1.4. The RunInfo.xml file was edited to allow for the 6 bp index to be called as a separate index read in the analysis, with matrix and phasing estimated from the PhiX control lane. This allowed the 6 bp index to be ignored in the alignment of the sequences to the reference genome (hg18) to facilitate calculation of basic QC measures, i.e. % clusters passing filters and alignment to hg18 (Supplementary Table SB). Both indexed and non-indexed sequence reads were treated in this way so that the effective read length for samples was 34 bp (for 40 bp reads) and 74 bp (for 80 bp reads). The sequenced reads were then parsed based on the index using an in-house Perl script to allow analysis of the data on an individual per sample basis. SNP detection and generation of data used to determine coverage were performed using MAQ13 on the individual sample sequence data. The Illumina quality scores were converted to the standard Phred scores required by MAQ by using a modified version of the fq_all2std.pl script supplied with MAQ. A SNP masked reference genome was used for alignment of the reads.
3. Results
Figure 1 outlines the experimental design for this study. Four sequencing libraries were enriched for 377 kb of target sequence. Libraries 1 and 2 each contained the same HapMap DNA sample but differed because library 2 included a 6 bp index. Comparison of sequence data from these indexed and non-indexed samples determined the impact of the index on the efficiency of the enrichment reaction. The third and fourth libraries were multiplex libraries containing three- and nine-indexed HapMap samples, respectively. Sequence data from these libraries were used to assess the performance of the indexes in distributing the reads from a single sequencing reaction to multiple samples and to measure the accuracy of SNP calling in multiplex samples.
3.1. Comparison of on-target versus off-target sequence coverage in indexed and non-indexed samples
Sequence data from the non-indexed DNA sample show that 20% of sequence reads were on-target, i.e. they mapped back to a target region of the enrichment reaction ±50 bp (Table 1). Across the target regions, there was a 1708-fold enrichment of target DNA in this sample. Ninty-eight percent of the targeted bases were covered by at least one sequence read and on average the target regions were covered to a depth of 169x. We investigated the effect of including a 6 bp index in the adaptor sequence on the efficiency of the enrichment reaction by enriching and sequencing the same DNA sample with an index. The on-target (22%) and fold enrichment (1885) metrics are similar for both samples (Table 1) indicating that the inclusion of the index did not compromise the performance of the enrichment reaction. Figure 2A and B show that for both ‘on-target’ and ‘off-target’ locations, the pattern of sequence coverage obtained is consistent between the single indexed and single non-indexed samples. The higher sequence coverage observed for the non-indexed sample compared with the indexed sample reflects the larger number of clusters (and therefore sequence reads) that passed QC filtering for the non-indexed sample (Supplementary Table SB). The three-index sample and the nine-index sample libraries had medians of 21 and 18% on-target sequence and medians of 1689- and 1467-fold enrichment, respectively, based on combined 40 and 80 bp read data. Individual sample level data and further quality control information are detailed in Supplementary Tables SB and SC.
Table 1.
Percentage on-target and fold enrichment for each library
| Non-index sample | One-index sample | Three-index samplea | Nine-index samplea | |
|---|---|---|---|---|
| Percentage reads in targeted regions ± 50 bp (%)b | 20 | 22 | 21 | 18 |
| Fold enrichment in targeted regionsc | 1708 | 1885 | 1689 | 1467 |
| Percentage target bases covered (%)d | 98 | 98 | 98 | 98 |
| Median coverage of targete | 169xf | 93xf | 164x | 46x |
aForty and 80 bp data were combined for the three-index and nine-index samples. Average values given for multisample libraries. Individual values are listed in Supplementary Table SB.
bNumber of reads uniquely mapping to the target region (±50 bp) as a % of the number of reads uniquely mapping to hg18.
c(Sequence reads uniquely mapping to the target regions/Sequence reads mapping to hg18) × Maximum enrichment where maximum enrichment is a ratio of genome length (3 080 419 510 bp) to target length (377 388 bp).
dPercentage of target bases covered by at least one sequence read.
e(Number of 34 or 74 bp reads matching target × 34 or 74)/target length.
fThe difference in median read coverage between the non-indexed and indexed sample is reflective of the larger number of clusters on the flowcell and also the larger number of clusters passing QC filters in the non-indexed sample (83.48 versus 57.65%, Supplementary Table SB).
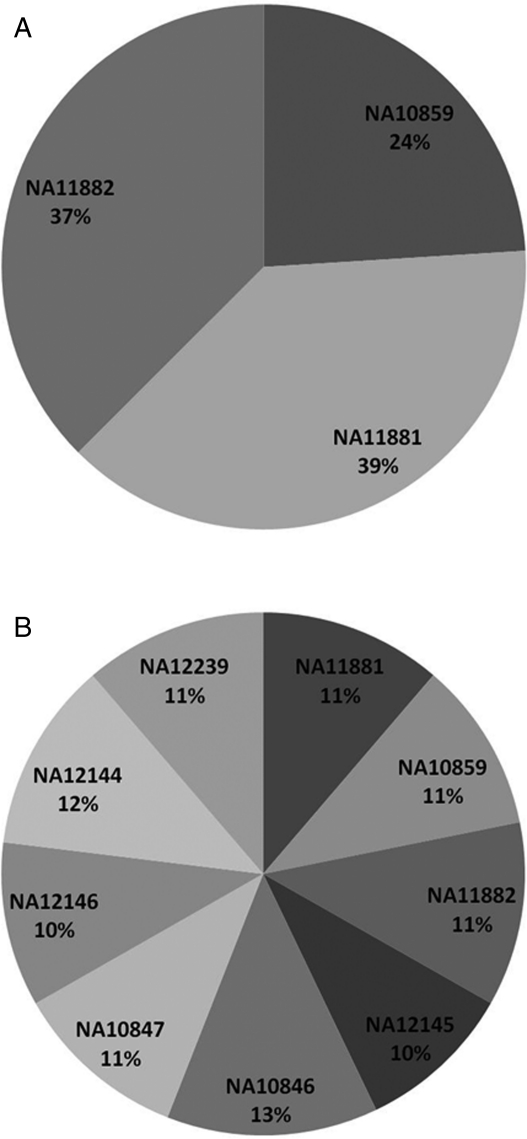
3.2. Comparison of read count for three-index sample versus nine-index sample
Figure 3 shows the performance of the indexing methodology in combination with target enrichment. The numbers of reads attributable to each index/sample are calculated as a percentage of the total number of sequence reads per lane of data. For both the three-index sample and nine-index sample libraries, there is a relatively even share of sequence reads for the individual samples, especially in the nine-index library. This indicates that the enrichment process has been consistent across all samples within each indexed library. The individual read counts uniquely aligning to the hg18 reference, percentage on-target and fold enrichment for each sample in the three- and nine-index sample libraries are detailed in Supplementary Table SC.
Figure 3.
Percentage of sequence reads per indexed sample in sequenced libraries. Percentage distribution per sample of sequence reads (pre-alignment to reference genome; 40 and 80 bp data combined) for the three-index (A) and the nine-index (B) sample libraries. The relative underperformance of sample NA10859 in the three-index library is not observed in the nine-index library and is unlikely to be due to a systemtaic problem with the ACACAT index.
3.3. Concordance of SNP calls with known HapMap genotypes
To illustrate the capacity of this method to detect SNPs with very high accuracy, we present data from the sequenced PTBP2 and CDC42 regions. Concordance rates for SNP calls compared with known HapMap genotypes are listed in Table 2. Only SNPs that had at least one copy of the non-reference allele, a sequencing depth of ≥8x and MAQ base quality score >30 were considered for analysis for each test sample. For the three-index sample library at a sequencing coverage of ≥8x, the concordance rate across the three samples was 99.1%. For the nine-index sample library at ≥8x coverage, the concordance rate was 98.9%. Combined these data give an overall concordance rate of 99% for SNP calls in the three- and nine-indexed samples. If we consider the concordance of SNP calls at all sites, the false-negative rate (i.e. the proportion of HapMap SNPs not detected in the sequence data irrespective of coverage) was 1.4% for the three-index sample library and 4.9% for the nine-index sample library. This assumes that the genotypes in the HapMap database are correct.
Table 2.
Concordance of SNPs called by MAQ in sequencing data with known HapMap genotypes
| Individual ID (# SNPs with at least one non-reference allele in PTBP2 and CDC42 target region for this sample) | Number of concordant SNP calls/number of SNPs with at least 8x coverage and Phred-like consensus quality > 30 (% concordance call) |
|
|---|---|---|
| Three-index samples | Nine-index samples | |
| NA11881 (103) | 102/103 (99.0%) | 96/97a (98.9%) |
| NA11882 (136) | 134/135 (99.3%) | 132/134 (98.5%) |
| NA10859 (106) | 104/105 (99.0%) | 98/99 (98.9%) |
| NA12144 (103) | 96/97 (98.9%) | |
| NA12239 (57) | 54/56 (96.4%) | |
| NA12145 (111) | 107/107 (100%) | |
| NA10846 (109) | 104/105 (99%) | |
| NA10847 (109) | 106/107 (99%) | |
| NA12146 (106) | 101/102 (99%) | |
aOnly 97 of the 103 SNPs had ≥8x coverage and a Phred-like consensus score >30 and were included in concordance analysis.
4. Discussion
We present a method where multiple DNA samples are first indexed and combined into one sequencing library and then target enriched using a single reaction. By indexing prior to enrichment, this method (i) dramatically reduces the costs associated with target enrichment and (ii) introduces significant flexibility into the design of targeted next-generation sequencing studies.
The efficiency of the enrichment reaction is not compromised by the inclusion of the 6 bp index in adaptor sequence as determined by enrichment and sequencing of the same DNA sample with and without an index. We were able to show that use of index-specific blocking oligos during the enrichment process is not necessary. The enrichment process is reasonably consistent across all indexed samples in both the three-index sample and nine-index sample libraries analyzed in this study. The problem of differential ligation efficiency for individual indexes11 has been resolved here by quantifying each ligated and PCR-enriched sequencing library and then combining individual indexed sample libraries in equimolar amounts prior to target enrichment. In the three-index sample library, the sample with the ACACAT index (NA10859) had approximately one-third less reads than the other two indexed samples in that library. However, in the nine-index sample library, where the read counts for all nine individual samples were within a tight range of each other, the same ACACAT index did not under perform compared with the other indexes. Differences in sample read counts are more likely due to pipetting or quantitation error when the sequencing libraries are initially combined prior to the enrichment process.
We demonstrate that multiplex target enrichment using DNA indexing can detect SNPs with high accuracy. In accordance with other next-generation sequencing studies, SNP detection is highly dependent on sequence coverage and is more difficult for heterozygous sites.14 Even at a relatively low coverage of ≥8x, 99% of SNPs were called correctly when data from the PTBP2 and CDC42 loci from the three- and nine-index samples were compared with online HapMap data. This concordance call rate compares well with rates reported for other target enrichment studies: 99.7% for variants with a sequencing coverage >5x, MAQ quality score >3014; 99.4% for high-quality calls inferred using Bayesian model12 and 99.57% for variants with a sequencing coverage >8x, MAQ quality score >30.15
Alternative methods amenable to combining multiple target enrichment and sample indexing such as high-throughput microdroplet-based PCR technology7 and on-array sequence capture8,15,16 require up to 7.5 and 20 µg of input DNA, respectively. These large requirements for input DNA put a significant strain on studies with limited amounts of source biological material. We used 3 µg of input DNA for our method but this could be lowered to 1 µg or less because the input requirement for the enrichment reaction is only 500 ng of Illumina sequencing library. Therefore, we believe our method is ideally suited to multiplex target enrichment of samples with limited DNA resources, e.g. clinical samples.
The factors that affect sequence coverage, and consequently SNP detection, are the volume of aligned sequence data generated from an experiment and the efficiency of the enrichment reaction. The former can be increased by repeating the sequencing experiment and/or loading a higher quantity of library DNA onto the flowcell to generate more data, sequencing to a longer read length, performing paired-end sequencing and implementing newer calling algorithms that can identify more sequence reads on a flowcell. The efficiency of the enrichment reaction is largely dependent on its design. In this study, we used a relaxed repeat masking protocol in an effort to increase the proportion of the PTBP2 and CDC42 genes that could be sequenced. Although this did allow us to sequence more of these genes than would have been possible with the stricter Agilent default repeat masking settings, it came at a cost of generating more off-target sequence. On-target sequence using this enrichment reaction was ∼21%. This is lower than achieved in other studies that used the SureSelect system (42–5012 and 40–45%14) or Agilent microarray capture (36–76,17 36–5516 and 37–54% for whole exome resequencing15). In this study, the reduction in on-target specificity is most likely due to the relaxed repeat masking used in the design of the target enrichment reaction, which resulted in a large proportion of reads mapping to repetitive regions (Supplementary Fig. SB). The advantage of implementing the default repeat masking settings during SureSelect eArray design will be greater on-target specificity. The disadvantage of these settings is that it will not be possible to target some genomic regions. How punitive this will be depends on the proportion of target regions that contains repetitive sequences. It will certainly impact on studies that plan to sequence entire genes including introns. For example, in this study, just under 40% of CDC42 could be targeted with default repeat masking settings. However, studies targeting exonic sequence at multiple sites will be less influenced by repetitive sequence.
In summary, we have designed an effective strategy to allow target enrichment and sequencing of multiple samples in parallel using DNA indexing. This method is very accurate for SNP detection. By combining DNA samples prior to enrichment, this method dramatically saves on enrichment and sequencing costs, and the inclusion of the DNA index permits each sample to be analyzed individually downstream. The method is an important inclusion in the range of next-generation sequencing applications below the level of whole exome resequencing but with the capability to sequence the exonic regions of hundreds of genes in tens of samples in a single sequencing experiment.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This material is based on works supported by the Science Foundation Ireland (SFI/07/RFP/GEN/F327/EC07) and the Health Research Board (Ireland) (HRB/HRA/2009/45).
References
- 1.Dahl F., Gullberg M., Stenberg J., Landegren U., Nilsson M. Multiplex amplification enabled by selective circularization of large sets of genomic DNA fragments. Nucleic Acids Res. 2005;33:e71. doi: 10.1093/nar/gni070. doi:10.1093/nar/gni070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landegren U., Schallmeiner E., Nilsson M., et al. Molecular tools for a molecular medicine: analyzing genes, transcripts and proteins using padlock and proximity probes. J. Mol. Recognit. 2004;17:194–7. doi: 10.1002/jmr.664. doi:10.1002/jmr.664. [DOI] [PubMed] [Google Scholar]
- 3.Li J.B., Gao Y., Aach J., et al. Multiplex padlock targeted sequencing reveals human hypermutable CpG variations. Genome Res. 2009;19:1606–15. doi: 10.1101/gr.092213.109. doi:10.1101/gr.092213.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lovett M., Kere J., Hinton L.M. Direct selection: a method for the isolation of cDNAs encoded by large genomic regions. Proc. Natl Acad. Sci. USA. 1991;88:9628–32. doi: 10.1073/pnas.88.21.9628. doi:10.1073/pnas.88.21.9628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Porreca G.J., Zhang K., Li J.B., et al. Multiplex amplification of large sets of human exons. Nat. Methods. 2007;4:931–6. doi: 10.1038/nmeth1110. doi:10.1038/nmeth1110. [DOI] [PubMed] [Google Scholar]
- 6.Rodriguez J.A., Guiteau J.J., Nazareth L., et al. Sequencing the full-length of the phosphatase and tensin homolog (PTEN) gene in hepatocellular carcinoma (HCC) using the 454 GS20 and Illumina GA DNA sequencing platforms. World J. Surg. 2009;33:647–52. doi: 10.1007/s00268-008-9852-x. doi:10.1007/s00268-008-9852-x. [DOI] [PubMed] [Google Scholar]
- 7.Tewhey R., Warner J.B., Nakano M., et al. Microdroplet-based PCR enrichment for large-scale targeted sequencing. Nat. Biotechnol. 2009b;27:1025–31. doi: 10.1038/nbt.1583. doi:10.1038/nbt.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Turner E.H., Lee C., Ng S.B., Nickerson D.A., Shendure J. Massively parallel exon capture and library-free resequencing across 16 genomes. Nat. Methods. 2009;6:315–6. doi: 10.1038/nmeth.f.248. doi:10.1038/nmeth.f.248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Varley K.E., Mitra R.D. Nested Patch PCR enables highly multiplexed mutation discovery in candidate genes. Genome Res. 2008;18:1844–50. doi: 10.1101/gr.078204.108. doi:10.1101/gr.078204.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mamanova L., Coffey A.J., Scott C.E., et al. Target-enrichment strategies for next-generation sequencing. Nat. Methods. 2010;7:111–8. doi: 10.1038/nmeth.1419. doi:10.1038/nmeth.1419. [DOI] [PubMed] [Google Scholar]
- 11.Craig D.W., Pearson J.V., Szelinger S., et al. Identification of genetic variants using bar-coded multiplexed sequencing. Nat. Methods. 2008;5:887–93. doi: 10.1038/nmeth.1251. doi:10.1038/nmeth.1251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gnirke A., Melnikov A., Maguire J., et al. Solution hybrid selection with ultra-long oligonucleotides for massively parallel targeted sequencing. Nat. Biotechnol. 2009;27:182–9. doi: 10.1038/nbt.1523. doi:10.1038/nbt.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Ruan J., Durbin R. Mapping short DNA sequencing reads and calling variants using mapping quality scores. Genome Res. 2008;18:1851–8. doi: 10.1101/gr.078212.108. doi:10.1101/gr.078212.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tewhey R., Nakano M., Wang X., et al. Enrichment of sequencing targets from the human genome by solution hybridization. Genome Biol. 2009;10:R116. doi: 10.1186/gb-2009-10-10-r116. doi:10.1186/gb-2009-10-10-r116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ng S.B., Turner E.H., Robertson P.D., et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–6. doi: 10.1038/nature08250. doi:10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hodges E., Xuan Z., Balija V., et al. Genome-wide in situ exon capture for selective resequencing. Nat. Genet. 2007;39:1522–7. doi: 10.1038/ng.2007.42. doi:10.1038/ng.2007.42. [DOI] [PubMed] [Google Scholar]
- 17.Albert T.J., Molla M.N., Muzny D.M., et al. Direct selection of human genomic loci by microarray hybridization. Nat. Methods. 2007;4:903–5. doi: 10.1038/nmeth1111. doi:10.1038/nmeth1111. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.