Abstract
The whole genome of Jatropha curcas was sequenced, using a combination of the conventional Sanger method and new-generation multiplex sequencing methods. Total length of the non-redundant sequences thus obtained was 285 858 490 bp consisting of 120 586 contigs and 29 831 singlets. They accounted for ∼95% of the gene-containing regions with the average G + C content was 34.3%. A total of 40 929 complete and partial structures of protein encoding genes have been deduced. Comparison with genes of other plant species indicated that 1529 (4%) of the putative protein-encoding genes are specific to the Euphorbiaceae family. A high degree of microsynteny was observed with the genome of castor bean and, to a lesser extent, with those of soybean and Arabidopsis thaliana. In parallel with genome sequencing, cDNAs derived from leaf and callus tissues were subjected to pyrosequencing, and a total of 21 225 unigene data have been generated. Polymorphism analysis using microsatellite markers developed from the genomic sequence data obtained was performed with 12 J. curcas lines collected from various parts of the world to estimate their genetic diversity. The genomic sequence and accompanying information presented here are expected to serve as valuable resources for the acceleration of fundamental and applied research with J. curcas, especially in the fields of environment-related research such as biofuel production. Further information on the genomic sequences and DNA markers is available at http://www.kazusa.or.jp/jatropha/.
Keywords: Jatropha curcas L., genome sequencing, cDNA sequencing, microsatellite markers
1. Introduction
To reconcile increasing energy consumption with worsening global environmental conditions is a fundamental concern of the contemporary society. Fossil fuel deposits are rapidly diminishing, and their consumption raises carbon dioxide discharge levels. Alternative fuels, such as bioethanol and biodiesel, show great promise for alleviating the problems caused by the consumption of fossil fuel.
Jatropha curcas L. is a plant belonging to the family Euphorbiaceae that is endemic to tropical America. It is now grown commercially in tropical and subtropical Africa and Asia. Jatropha has considerable potential for various uses including biofuels.1 The plant can grow at rainfall levels as low as 200 mm per annum.2 Medicinal compounds are found in various parts of the plant,1 but it is the potentially high yield of oil per unit land area, which is second only to oil palm,3 that makes Jatropha an outstanding biofuel plant. Furthermore, the quality of oil in its seeds is suitable for production of biodiesel as they contain more than 75% unsaturated fatty acids.4 Despite its cultivation throughout the tropical and subtropical world, the positive attributes of this plant are not fully understood in terms of breeding and utilization.3 This can be attributed mainly to the lack of information on its genetics and genomics. The genome size (∼410 Mb) and the base composition have been estimated by flow cytometry, and karyotypes have been characterized.5 Expressed sequence tags (ESTs) from developing and germinating Jatropha seeds have been reported.6 However, no further information on the genomic structure of J. curcas is available.
To understand the genetic system of this plant and to accelerate the process of molecular breeding, we analysed the structure of the whole genome of J. curcas. For genome sequencing, we adopted a combination of BAC end sequencing and shotgun sequencing by the conventional Sanger method and the new-generation multiplex methods, which was followed by information analyses. In addition, microsatellite markers have been developed using the sequence information, and polymorphism among various J. curcas varieties was examined. The information and material resources for the Jatropha genome generated in this study will enhance both fundamental and applied research with J. curcas and related plants.
2. Materials and methods
2.1. Plant materials
A J. curcas line originating from the Palawan Island in the Philippines was subjected to genome sequencing. The following 12 lines were used for diversity analysis: Palawan, Indonesia, Indonesia IS, Thai, Chinese, Mexico 2b, Guatemala 1, Guatemala 2, Tanzania, Madagascar, Cape Verde, and Uganda. The Indonesia IS and Thai lines were purchased from IS Co. Ltd. (Tokyo, Japan) and Nikko-Seed Co. Ltd. (Tochigi, Japan), respectively. The Uganda and the remaining nine lines were kindly provided by BBL International (Osaka, Japan) and Nippon Biodiesel Fuel Co., Ltd. (Tokyo, Japan), respectively.
2.2. Construction of BAC libraries
BAC genomic libraries were constructed using the genomic DNA of J. curcas partially digested with either MboI or HindIII and Copy Control pCC1BAC as a cloning vector. The average insert size of these libraries was 80.2 kb for the MboI library and 94.9 and 63.4 kb for two independent preparations of the HindIII libraries. Both libraries covered the haploid genome 9.2 times in total.
2.3. BAC sequencing
To analyse end sequences, BAC DNAs were amplified using a TempliPhi large construction kit (GE Healthcare, UK), and the end sequences were analysed according to the Sanger method using a cycle sequencing kit (Big Dye-terminator kit, Applied Biosystems, USA) with DNA sequencer type 3730xl (Applied Biosystems). High-quality BAC sequences were determined by the shotgun method using the Sanger sequencing protocol, as described previously.7
2.4. Shotgun genomic sequencing
For sequencing by the Sanger method, shotgun libraries with average insert sizes of 2.5 kb were generated using pBluescript SK- as a cloning vector, and these were used to transform Escherichia coli ElectroTen-Blue (Agilent Technologies, Santa Clara, CA, USA). The shotgun clones were propagated in microtiter plates, and the plasmid DNA was amplified using a TempliPhi kit (GE Healthcare). Sequencing was performed using a cycle sequencing kit (Big Dye-terminator Cycle Sequencing kit, Applied Biosystems) with DNA sequencer type 3730xl (Applied Biosystems) or DeNOVA-5000HT (Shimadzu Co., Japan) according to the protocols recommended by the manufacturers.
High-throughput multiplex sequencing was carried out using a Genome Sequencer (GS) FLX Instrument (Roche Diagnostics, USA) and Genome Analyzer II (Illumina Inc., USA) sequencers. A 5-µg sample of Jatropha total cellular DNA was sheared by nebulization and subjected to library preparation followed by shotgun sequencing using the GS FLX platform. For the 3-kb paired-end sequencing, the library was prepared using GS Titanium Library Paired End Adaptors according to the manufacturer's instructions. For sequencing by an Illumina-solexa GAII sequencer, the sample was prepared according to the manufacturer's manual. Briefly, 1 µg of the total cellular genomic DNA was fragmented by the Covaris S1 instrument (Covaris Inc.). The fragmented DNA was repaired, and the adapters for paired-end sequencing (36, 51, and 76 cycles) were then ligated to the repaired DNA fragment. The size-selected fragment (300–350 bp) by agarose gel electrophoresis was PCR amplified, and the PCR product was validated using a 2100 Bioanalyzer (Agilent Technologies) and a 7900HT Fast Real-Time PCR system (ABI). The sample was then run on a Genome Analyzer II using the 36 cycles sequencing kits. Base-calling was performed using the Genome Analyzer Pipeline.
2.5. cDNA sequencing
Total RNA was extracted from leaf and callus tissue using an RNeasy Plant Mini Kit (Qiagen, Germany). mRNA was purified from the total RNA using Oligotex-dT30 (Takara Bio Inc., Japan). Sequencing was performed with a GS FLX Instrument (Roche Diagnostics) using the cDNA rapid library method according to the manufacturer's instructions.
2.6. Assembly of sequence data
Reconstruction of the genome sequence of J. curcas was performed in the following two steps: assembly of sequence data generated by different types of DNA sequencers, and scaffolding and base correction.
The sequence data collected according to the Sanger protocol using a 3730xl capillary sequencer were subjected to trimming of sequences derived from cloning vectors with the Figaro and Lucy programs,8 followed by assembly with the PCAP.rep program.9 Base-calling of the sequence data generated by pyrosequencing using a GS FLX sequencer was performed using the Pyrobayes program.10 The sequence reads artificially replicated during an emulsion PCR were removed by a 454 replicate filter,11 and the remaining reads were assembled using MIRA version 3 rc4 software.12 Contigs and singlets generated by assembly using the Sanger protocol and pyrosequencing were separately subjected to similarity searches for sequences of the chloroplast (GenBank: FJ695500) of J. curcas and the mitochondria (GenBank: Y08501) of Arabidopsis thaliana13 using the Megablast program.14 Matching sequences were then removed. All remaining contigs and singlets were assembled using the PCAP.rep program.9 BAC end sequences in which the vector sequences were trimmed by Figaro and Lucy programs8 in advance were further integrated in the resulting sequences using PCAP.rep.9 Then, sequences 99 bp and shorter were removed.
The resulting contigs and singlets were designated as follows. The contigs containing sequences from the Sanger sequencing, pyrosequencing, and BAC end sequencing were prefixed with ‘JcCA’ followed by a seven-digit number. The contigs containing sequences from both the Sanger and 454 sequencing were prefixed with ‘JcCB’ followed by a seven-digit number. The contigs containing sequences from the Sanger sequencing and the pyrosequencing were prefixed with ‘JcCC’ and ‘JcCD', respectively. The singlets from the Sanger sequencing and pyrosequencing that were not assembled into other sequences throughout the whole process were prefixed with ‘JcSR’ and ‘JcPR', respectively.
For improvement of data quality, both single and mate-pair reads by an Illumina GAII sequencer were collected and assembled using the Velvet program.15 The resulting contig sequences were mapped onto the contigs generated by hybrid assembly to correct the short insertion–deletion (indels) errors.
Both paired-end reads of the genomic DNA and single reads of cDNAs by the GS FLX sequencer were used for scaffolding. Paired-end reads of the genomic DNA were assembled with the MIRA program12 according to the manufacturer's instructions, and the resulting sequences were used for scaffolding of the contig sequences generated by the Sanger sequencing and pyrosequencing using the GS reference mapper ver. 2.3 program. In parallel, a mixture of cDNAs derived from leaf and callus tissue of Jatropha was subjected to sequencing by GS FLX, and the data obtained were subjected to assembly with the MIRA ver. 3.0.5 program in the EST mode.12 In addition, the resulting cDNA sequences as well as the Jatropha ESTs retrieved from public DNA databases were used for scaffolding using the Blat program.16
2.7. Gene assignment
Gene prediction and modelling were performed by automatic gene assignment programs that employ ab initio gene finding and similarity searches. For ab initio gene finding, predictions of protein-coding regions were carried out using GeneMark.hmm17 and Genescan18 programs with the matrix trained by an A. thaliana gene set, and predictions of exon–intron structure were performed using NetGene219 and SplicePredictor20 programs. Similarity searches for potential protein-coding regions and all contigs were performed against a Uniref database (http://www.ebi.ac.uk/uniref/) using Blastp and Blastx programs21 with a cut-off (E-value ≤ 1e−3). The exon–intron structure of potential protein-coding regions and the contigs homologous to the Uniref database (http://www.ebi.ac.uk/uniref/) were predicted using the Nap program.22 Suitable exon-intron structures were determined by considering all the information above. The predicted gene structures were further confirmed by comparison to cDNA sequences analysed in this study. The protein-coding genes assigned in this manner were denoted by IDs with the contig names followed by sequential numbers from one end to another. They were classified into four categories based on sequence similarity to registered genes: genes with complete structure, pseudogenes, genes with partial structure, and transposons/retrotranspons.
2.8. Functional assignment and classification of potential protein-coding genes
To assign the gene families, functional domains, GO terms, and GO accession numbers,23 the predicted genes were searched against InterPro using InterProScan24 software. Genes with an E-value of <1.0 were taken into account. GO terms were grouped into plant GO slim categories using the map2slim program (http://www.geneontology.org/GO.slims.shtml).
The predicted protein-encoding genes were mapped onto KEGG metabolic pathways25 using the Blastp program21 against the GENES database.25 Thresholds of amino acid sequence identity ≥25% and of length coverage of the query sequence ≥50% with a cut-off (E-value ≤ 1e−10) were applied.
2.9. Phylogenetic analysis
Evolutionary relationships of proteins of casben synthase genes, disease resistance genes, MADS-box genes, flowering genes, and COL genes were analysed using predicted amino acid sequences from different databases aligned with the program CLUSTALW (Ver. 1.83).26 Evolutionary relationships were inferred using a neighbour-joining algorithm.27 All positions containing alignment gaps and missing data were eliminated only in pairwise sequence comparisons. Phylogenetic trees were constructed with MEGA4 software.28
2.10. Polymorphism analysis
Microsatellite or simple sequence repeats (SSRs) 15 nucleotides in length, containing all possible combinations of di-nucleotide (NN), tri-nucleotide (NNN) and tetra-nucleotide (NNNN) repeat, were identified from the Jatropha genome sequences using the SSRIT (SSR Identification Tool) program.29 Primer pairs for amplification of SSR-containing regions were designed based on the flanking sequences of each SSR with the Primer 3 program30 so that amplified fragment sizes were between 90 and 300 bp in length. One hundred microsatellite markers were subjected to examination of polymorphisms among 12 lines of J. curcas.
PCR amplifications (5 µl) were performed on 0.7 ng of Jatropha genomic DNA in 1 × PCR buffer (BIOLINE, London, UK), 3 mM MgCl2, 0.04 U BIOTAQ™ DNA Polymerase (BIOLINE), 0.8 mM dNTPs, and 0.4 μM of each primer, using the modified ‘Touchdown PCR’ protocol described by Sato et al.7 PCR products were separated by 10% polyacrylamide gel electrophoresis using TBE buffer, and data were collected as described previously. Allele detection and genotype code typing were performed using the Polyans program (ver.1.1; http://www.kazusa.or.jp/polyans). The presence or the absence of amplification and the number of different-sized fragments, which was taken as the number of alleles, were recorded. Loci for which there was no amplification were designated as null alleles. PIC was calculated using the following equation:
where Pij is the frequency of the jth allele for the ith locus. NTSYSpc ver. 2.21c software (Applied Biostatistics Inc., New York, USA) was employed to perform cluster analysis. The SimQual and SAHN modules were used for estimation of genetic distance and a genetic tree, respectively, with the coefficient in SimQual set to SM, and the clustering method set to UPGMA.
3. Results and discussion
3.1. Sequence analysis of the Jatropha genome
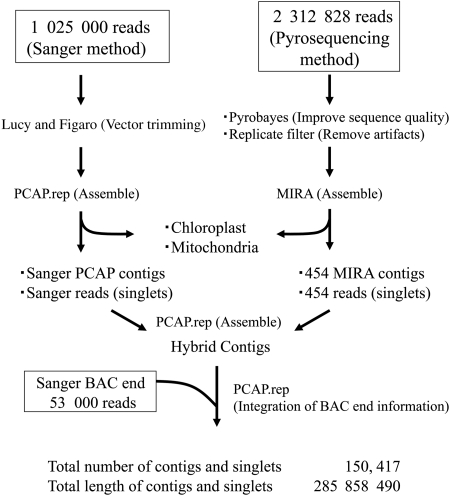
The strategy and the status of sequencing and assembly are summarized in Fig. 1. Briefly, the 1 025 000 reads of the Sanger sequencing and the 2 312 828 reads of pyrosequencing, which were appropriately processed in advance as indicated in Fig. 1, were independently assembled using the PCAP.rep9 and MIRA programs,12 respectively. The resulting contigs and singlets were subjected to hybrid assembly by PCAP.rep,9 and the 53 000 BAC end sequences were further integrated.
Figure 1.
The strategy and status of sequencing and assembly.
For improvement of data quality, 86 028 428 (36 bases long from each end for each read) and 96 580 336 short-reads (50 and 31 bases long from each end for each read) by mate-pair sequencing with the Illumina GAII sequencer were assembled into 569 576 contigs (total length: 75 539 079 bp) by the Velvet program.15 The resulting contig sequences were mapped onto those generated by hybrid assembly to correct short indels errors. These indels were probably attributed to classified insertions, deletions, and mismatches by their association with miscall from homopolymer effects. As a result of mapping, 7459 loci on the 5025 contigs were revised.
A total of 695 928 3-kb paired-end reads by the GS FLX sequencer were used for scaffolding of the generated contigs and singlets using the GS reference mapper ver. 2.3 program, as described in the ‘Materials and Methods’ section. In parallel, 991 050 reads of cDNA sequences by pyrosequencing were collected; 534 137 were derived from leaf tissue and 456 913 from callus tissue. The cDNA sequences were assembled with MIRA 3.05 in the EST mode,12 and 21 225 unigene sets were generated consisting of 13 610 contigs and 7615 singlets used for scaffolding by the BLAT program.16 In addition, unigenes generated from 26 447 ESTs registered in public DNA databases (http://www.ncbi.nlm.nih.gov/dbEST/) were also used for scaffolding. As a result of scaffolding, the 44 153 contigs and singlets constructed by hybrid assembly were integrated into 15 300 scaffolds. The total length of the scaffolds was 129 291 074 bp. The longest scaffold (JcS_100001) had 56 042 bp, and the average scaffold length was 8450 bp. The constructed scaffolds were designated as JcS followed by sequential numbers.
The total length of the final genomic sequences of J. curcas obtained was 285 858 490 bp, consisting of 120 586 contigs (276 710 623 bp total) and 29 831 singlets (9 147 867 bp total), which is ∼70 and 75% of the whole genome of 4105 and 380 Mb (N. Wada, unpublished result), respectively, estimated by flow cytometry. The average length of contigs and singlets was 1900 bp. Statistics of the assembly are summarized in Table 1. The longest contig was 29 744 bp, and N50 length was 3833 bp. The distribution of contig lengths is shown in the Supplementary Fig. S1. The average G + C content of the contigs was 34.3%.
Table 1.
Assembly statistics
| Total length of contigs and singlets | 285 858 490 |
| Total number of contigs and singlets | 150 417 |
| Average length of contigs and singlets | 1900 |
| Maximum length of contigs and singlets | 29 744 |
| N50 | 3833 |
| G + C content (%) | 34.3 |
| Number of contigs (JcCA) | 32 212 |
| Number of contigs (JcCB) | 60 363 |
| Number of contigs (JcCC) | 2483 |
| Number of contigs (JcCD) | 25 528 |
| Number of singlets (JcSR) | 26 819 |
| Number of singlets (JcPR) | 1347 |
| Number of BAC end sequences (JHL/JHS/JMS) | 1665 |
| Contigs | |
| Total number of contigs | 120 586 |
| Total length of contigs | 276 710 623 |
| Average length of contigs | 2295 |
| Singlets | |
| Total number of singlets | 29 831 |
| Total length of singlets | 9 147 867 |
| Average length of singlets | 307 |
Coverage of gene space in the Jatropha genomic sequences was estimated roughly by surveying the matched non-redundant cDNA sequences obtained in this study. Of 21 225 non-redundant cDNA sequences and 26 447 EST sequences in the public databases, 45 029 matched Jatropha genomic sequences with an identity of 95% or more for a stretch of 50 nucleotides, suggesting that 95% of the gene space in the Jatropha genome was covered by the genomic sequences in this study.
We adopted here the sequencing strategy that combines the conventional Sanger method and the new-generation multiplex sequencing methods with the aid of various computer software for assembly. This strategy is superior in that shortcomings of respective methods are compensated by each other, enabling acquisition of sequences of higher quality in lower cost within a shorter period of time, thus is becoming popular for genome sequencing in both bacteria and eukaryotes.
3.2. Characteristic features of the genome
3.2.1. Repetitive sequences
A total of 41 428 di-, tri-, and tetra-nucleotide SSRs ≥15 bp were identified in the Jatropha genomic sequences (Supplementary Table S1). The frequency of the occurrence of these SSRs was estimated to be one SSR in every 7.0 kb in the 289 Mb sequences of the Jatropha genome. The di-, tri-, and tetra-nucleotide SSRs accounted for 46.3, 34.3, and 19.4% of the identified SSRs, respectively (Supplementary Table S1). The SSR patterns that appeared frequently were (AT)n, (AAT)n, and (AAAT)n, each representing 71% of di-nucleotide, 60% of tri-nucleotide, and 58% of tetra-nucleotide repeat units, respectively. The tri-nucleotide SSRs, particularly (AAG)n and (AGC)n, were preferentially found in exons. (AT)n, (AG)n, and (AAT)n were enriched in 5′ and 3′ untranslated regions, and (AC)n frequently occurred in introns (Supplementary Table S1).
A search of the Jatropha genomic sequences using the repeat sequence finding program RECON31 unravelled the occurrence of a variety of repeat elements including class I and class II transposable element (TE) subfamilies and some that were difficult to classify into known subfamilies. Composition of these repeat sequences was analysed with the RepeatMasker program (http://repeatmasker.org/); the results are summarized in Table 2. The identified repetitive sequences in total occupied 36.6% of the Jatropha genomic sequences. The most abundant repeat category was class I TE (29.9%), in which Gypsy type (19.6%) and Copia type (8.0%) LTR retroelements constituted major components.
Table 2.
Repetitive sequences in the Jatropha genomic sequences
| Repeat type | Jatropha genomic sequences |
||
|---|---|---|---|
| Number of elements | Coverage (kb) | Percentage of sequence | |
| Class I | |||
| LINEs | 195 | 136.9 | 0.05 |
| LTR: Copia | 31 740 | 22 318.2 | 8.03 |
| LTR: Gypsy | 67 658 | 56 655.7 | 19.60 |
| LTR: other | 13 454 | 6436.6 | 2.23 |
| Total class I | 113 047 | 86 447.4 | 29.91 |
| Class II | |||
| Coding class II | 5709 | 4102.9 | 1.42 |
| MITE | 5980 | 1802.8 | 0.62 |
| Total class II | 11 689 | 5905.7 | 2.04 |
| Short tandem repeats | 2092 | 148.1 | 0.05 |
| Unclassified | 25 977 | 14 953.3 | 5.17 |
3.2.2. RNA-coding genes
A combination of computer prediction and similarity searches of the structural RNA sequence library resulted in identification of 597 putative genes for transfer RNAs in the Jatropha genomic sequences. Although 80 of these were likely to be pseudogenes, the remaining 517 could code for intact tRNAs with 54 species of anticodons (Supplementary Table S2). This is sufficient for translation of all the amino acids based on the universal codon table.
A total of 65 genes for snRNAs were assigned by referring to the list of A. thaliana snRNAs (Supplementary Table S3).32 Some of these genes were found on the same contigs and scaffolds; thus, they are likely to form clusters in the genome, as they do in A. thaliana.
3.3. Characteristic features of protein-encoding genes
3.3.1. Prediction of protein-encoding genes
The Jatropha genomic sequences were subjected to an automatic assignment of protein-encoding genes, and a total of 40 929 genes, besides 16 447 transposon-related genes, were assigned. Complete structures were predicted for 9870 genes, but only partial structures were predicted for 17 863 genes. In addition, 1960 and 11 236 genes were likely to be pseudogenes with complete and truncated structures, respectively. Of the 40 929 presumptive protein-encoding genes, 15 573 (38.0%) carried ESTs with sequence identity of 95% or more for a stretch of 50 nucleotides.
Structural features of the protein-encoding genes in J. curcas were investigated in detail for 146 genes predicted on the 17 BAC clones (1.36 Mb in total) for which high-quality sequences were obtained by manual finishing and annotation (Supplementary Table S4). As shown in Supplementary Table S5, the basic structures of the protein-encoding genes in J. curcas are similar to those of A. thaliana except for the average lengths of genes and introns: 3064 versus 1918 bp and 356 versus 157 bp in J. curcas and A. thaliana, respectively.
3.3.2. Gene components
A similarity search of translated amino acid sequences of the 40 929 presumptive protein-encoding genes was performed using the TrEMBL database as a protein sequence library.33 The results indicated that 31 822 (77.7%) genes had significant (E-value ≤ 1e−20) sequence similarity to those in this database. Of these genes, 13 067 (41.0%) genes showed sequence similarities to those in a public EST database (http://www.ncbi.nlm.nih.gov/dbEST/) with a cut-off (E-value ≤ 1e−20) using tBLASTN.
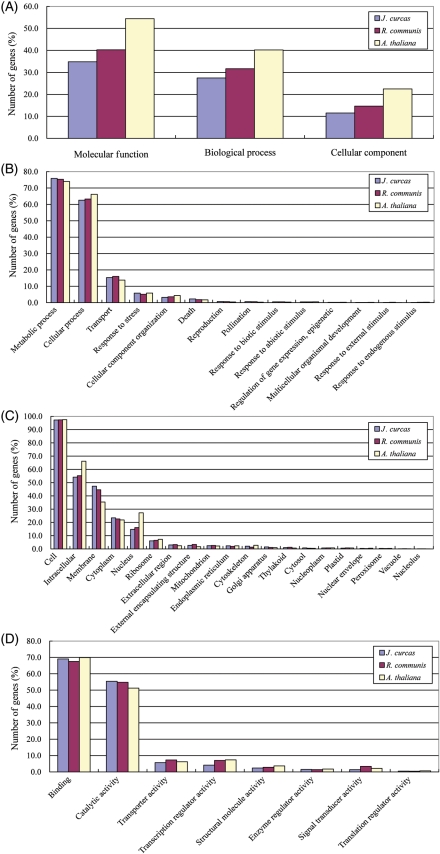
The 40 929 presumptive protein-encoding genes assigned in J. curcas and those in castor bean (Ricinus communis; 31 221 genes),34 which belongs to the same family as Jatropha, and A. thaliana (32 615 genes), were classified into plant GO slim categories35 for comparison (Fig. 2). The percentage of the number of genes classified into each GO slim category (i.e. ‘biological process’, ‘cellular component', and ‘molecular function’) was calculated for J. curcas, R. communis, and A. thaliana (Fig. 2).
Figure 2.
GO category classification. The percentages of number of genes classified into each GO slim category in J. curcas, R. communis, and A. thaliana are, respectively, shown in blue, red, and yellow bars. (A) GO terms; (B) biological process; (C) cellular component; and (D) molecular function.
Of 40 929 presumptive genes in the Jatropha genomic sequences, 2213 genes could be mapped onto 134 of the 155 metabolic pathways in the KEGG database,25 whereas the 2975 and 4115 genes of R. communis and A. thaliana were mapped onto 140 and 135 pathways, respectively. Twenty-nine pathways, including ‘fatty acid metabolism’ in lipid metabolism, ‘methionine metabolism’ and ‘lysine degradation’ in amino acid metabolism, and ‘benzoate degradation via hydroxylation’ in xenobiotics biodegradation and metabolism, contained enzyme(s) on which the genes in the Jatropha genome were solely mapped (Supplementary Table S6).
3.4. Characteristic features of the genes in J. curcas
3.4.1. Genes involved in synthesis of triacylglycerols
Jatropha curcas is expected to contribute to biodiesel production through its ability to biosynthesize and accumulate considerable amounts of triacylglycerols (TAGs) in seeds. For this reason, the genes involved in TAG biosynthesis are of great interest and some of those genes have already been cloned from J. curcas.36,37 Recently, the collection of ESTs from developing and germinating Jatropha seeds has been reported.6 We manually annotated and summarized the gene models for fatty acid and TAG biosynthesis that were predicted in this work, together with related data that have been deposited to GenBank (Supplementary Table S7). The Jatropha genome appears to contain basically one gene for each enzyme isoform, and no obvious gene duplication particular to this plant was identified in this category. One gene model for a recently identified soluble type of DGAT38 also existed in the Jatropha genome. To improve Jatropha oil quality for biodiesel, its fatty acid composition could be changed by altering the expression of some of the genes listed in Supplementary Table S7.
3.4.2. Genes related to phorbol ester biosynthesis
Jatropha curcas is known to produce tumour-promoting phorbol esters.39 Accordingly, depression of the phorbol ester biosynthetic gene in high oil content lines would be a step towards safe utilization of this plant. To our knowledge, genes involved in biosynthesis of phorbol esters have not been reported in J. curcas, with the exception of the gene for geranylgeranyl diphosphate synthase (GGPPS).40 In the current study, we searched genes for GGPPS, casbene synthase (CS), terpene hydroxylase (cytochrome P450-dependent monooxygenase), and acyltransferase in the Jatropha genome with the tBLASTN program21 using the corresponding amino acid sequences in diterpene-producing plants as queries (Supplementary Table S8).
One (JcCS1), two (JcCS2 and JcCS3), and six (JcCS4–JcCS9) homologues of a gene for CS in R. communis were identified in the BAC clones, JHL23C09, JHL22C18, and JHL17M24, respectively. JcCS2 is a pseudogene because there are several stop codons in the putative open reading frame (ORF). Interestingly, JcCS4–JcCS9 are tandemly aligned and are likely to be active because their ORFs seem to be intact. The phylogenetic tree demonstrates that JcCS4–JcCS9 forms a cluster, suggesting that continuous duplication of the original JcCS gene occurred recently (Supplementary Fig. S2 and Supplementary Table S9). There are 40 genes for terpenoid synthase (AtTPS) in A. thaliana that are most closely related to JcCS phylogenetically.41 They form clusters consisting of two or three tandem repeats at six loci in the genome. The clustered organization of JcCS may be an implication of the evolutionary process of genes related to the synthesis of terpenoid natural products.
3.4.3. Genes encoding curcin
Curcin is a Type I ribosome-inactivating protein (RIP) common among the members of the Euphobiaceae family. Curcin in J. curcas is analogous to ricin, a Type II RIP, in R. communis, although the toxicity of curcin is significantly lower than that of ricin.42 Research on curcin has been extensive,42 and it has revealed antitumour activity.43,44 The activity of a curcin protein isoform against viral and fungal diseases has been proven by heterologous expression in tobacco; the expression of this curcin gene was induced by abiotic and biotic stresses in leaves.45–47 So far, Jatropha genes encoding three isoforms of curcin have been reported and deposited in public DNA databases. In our Jatropha genome sequence, only three contigs were identified to encode amino acid sequences highly similar to those coding for curcin, confirming that the Jatropha genome contains three curcin genes. However, there are four more contigs with presumptive genes predicted to encode curcin-like proteins with E-values from 1e−117 to 1e−91, as listed in Supplementary Table S10, suggesting that at least two more curcin isoforms are encoded in the Jatropha genome because these four additional genes make two pairs with highly similar counterparts. Data from proteomic analysis of developing seeds that is briefly mentioned in Costa et al.6 appear to support this observation as they identified five isoforms of curcin.
3.4.4. Disease resistance genes
In response to pathogens, plants have evolved disease resistance (R) genes. Most of them are NBS-LRR (nucleotide-binding site and leucine-rich repeat) proteins, which are classified into two groups on the basis of the presence of Toll and human interleukin receptors (TIR) at their amino termini.48 We identified 42 TIR NBS-LRR proteins and 50 non-TIR NBS-LRR proteins. We analysed five BAC clones (JHL06P13, JHS03A10, JHL25H03, JHL25P11, and JMS10C05) including R genes to reveal their gene structure (Supplementary Table S4). Two BAC clones (JHS03A10 and JMS10C05) include singletons of JcTIR-NBS-LRR1 and JcNBS-LRR9, whereas three clones (JHL06P13, JHL25H03, and JHL25P11) contain gene clusters as tandem repeats of R genes, JcNBS-LRR1 and JcNBS-LRR2, JcNBS-LRR3–5, or JcNBS-LRR6–8. JcNBS-LRR8 is a pseudogene with a stop codon in the ORF. The phylogenetic tree of R genes including eight R genes in J. curcas demonstrated that JcNBS-LRR3–5 or JcNBS-LRR6 and JcNBS-LRR7 are closely related, suggesting that these gene clusters evolved recently by the way of gene duplication (Supplementary Fig. S3 and Supplementary Table S11). Interestingly, JcNBS-LRR1 and JcNBS-LRR2 belong to different clades. This relationship indicates that gene duplication was not recent and that these gene segments were conserved after evolutionary diversification of J. curcas.
3.4.5. MADS-box genes
MADS-box genes, typical homeotic genes coding for transcription factors, form a family and are involved in several aspects of plant development.49 Many plant species are known to harbour multiple MADS-box genes that belong to a range of functionally divergent subfamilies.50 We searched for MIKC type II MADS-box genes in the genome of J. curcas using amino acid sequences of PI in A. thaliana as a query. A total of 28 potential MADS-box genes (JcMADS01–JcMADS28) were identified (Supplementary Table S12). The phylogenetic analysis classified these genes into several subfamilies (Supplementary Fig. S4).
SVP controls flowering time by negatively regulating the expression of a floral integrator, FLOWERING LOCUS T in response to ambient temperature changes in A. thaliana.51 Interestingly, there are five paralogs of SVP in Jatropha, yet only a single copy and three copies were identified in A. thaliana and Oryza sativa, respectively.52,53 Eight paralogs of SVP copies have been found in 57 MIKC type II MADS-box genes of Populus trichocarta,54 suggesting amplification and functional diversification of the SVP gene in woody plants.
3.4.6. Flowering-related genes
Flowering in J. curcas is closely related to the production of seeds. Jatropha curcas is a monoecious species, which forms unisexual flowers, male and female flowers, separately in an individual plant. The unisexual flowers are produced on the same inflorescence, with the ratio of male flowers to female flowers ranging from 10:1 to 30:1.55 The male bias ratio within an inflorescence limits seed production because more female flowers mean more fruits. Accordingly, modification of floral identity genes involved in organ identity could change the number or size of male and female organs or flowers.
In the Jatropha genomic sequences, we identified eight orthologs of flowering-related genes including five flowering regulators, CONSTANS, FLOWERING LOCUS D, FLOWERING LOCUS F, LEAFY, and SUPPRESSOR OF OVEREXPRESSION OF CONSTANS 1, designated as JcCO, JcFD, JcFT, JcLFY, and JcSOC1, respectively, and three floral identity genes, APETALA2, APETALA3, and PISTILLATA, designated as JcAP2, JcAP3-1, and JcPI, respectively (Supplementary Table S13). Phylogenetic analysis indicated that all Jatropha flowering-related genes except JcCO are closely related to those of woody plants, including Betula pendula, Hevea brasiliensis, R. communis, and Vitis vinifera (Supplementary Fig. S5 and Supplementary Table S14). JcCO belonged to evolutionary lineages that differ from its homologues in monocot and dicot species. Further phylogenetic analysis indicated that JcCO is not related to any flowering-related genes including CO paralogs, a rice CO orthologue Hd1 and light-signalling genes of AtCOLs, which are CO-like genes in A. thaliana (Supplementary Fig. S6 and Supplementary Table S15). This finding suggests that JcCO is not directly involved in flowering regulation, although JcCO has all CO-conservative domains as a transcription factor including B-box and CCT motif. There were other CO homologues in the Jatropha genome; for example, JcCOL2 in JcCB0217351.10 and JcCOL9 in JcCA0317951.10, which suggests that different components participate in the response to light in J. curcas.
3.5. Comparative analysis
3.5.1. Genes conserved in the Euphorbiaceae
To identify genes conserved specifically in the family Euphorbiaceae, amino acid sequences translated from the putative Jatropha genes predicted in this study were compared with those of genes in the genomes of A. thaliana, O. sativa, P. trichocarpa, V. vinifera, L. japonicus, and Glycine max, as well as protein sequences in the TrEMBL protein database.33 Sequences from the predicted genes in the R. communis genome34 and the gene index database for cassava (Manihot esculenta) were used as references for Euphorbiaceae protein-encoding genes. BLAST searches with a cut-off (E-value ≤ 1e−20) indicated that 1529 genes (4% of the predicted protein-encoding genes) were found only in the Euphorbiaceae. The InterPro annotations of these Euphorbiaceae-specific genes were surveyed to find conserved motifs in these genes, and consequently, 22 InterPro motifs were likely to be conserved in five or more genes (Supplementary Table S16). Of these, the C1-like motif (IPR011424), the pentatricopeptide repeat motif (IPR002885), and the cytochrome P450 motif (IPR001128) were found in 10, 10, and 9 genes, respectively.
Furthermore, 1176 of the genes predicted in the Jatropha genome assembly had matching sequences only in the Jatropha cDNA database suggesting that these genes are specific to J. curcas. The most common InterPro motifs found in these genes were the protein kinase-like domain (IPR011009) detected in six genes (Supplementary Table S17). The entire list of the Euphorbiaceae- and Jatropha-specific genes is provided in Supplementary Tables S18 and S19, respectively.
3.5.2. Microsynteny
To investigate the syntenic relations between the Jatropha and the other plant genomes, status of conservation of relative gene positions was surveyed using the scaffolds of Jatropha genomic sequences. Among the 1556 scaffolds with five or more predicted genes, conservation of the relative positions of three or more genes was observed in 829 scaffolds (53%) against genes predicted in the R. communis genomic sequences34 (Supplementary Tables S20 and S21). It appears that a significant degree of synteny can be expected within the family Euphorbiaceae. A syntenic relationship was also detected against the genomes of G. max and A. thaliana to a lesser degree. Microsyntenic relations have been observed in 178 (11%) and 256 (16%) of the 1556 scaffolds of the Jatropha genomic sequences, respectively (Supplementary Tables S20 and S21). The microsyntenic relationships between these plant species may provide useful information for predicting gene organization in the ancestral genome of dicots.
3.5.3. Genetic diversity among Jatropha lines
Five SSR motives were found in the 100 genome-derived microsatellite markers tested. Most of the SSRs were poly (AT)n (83 SSRs), followed by poly (AAT)n (8 SSRs), poly (AG)n (5 SSRs), poly (AAG)n (3 SSRs), and poly (AC)n (1 SSR). A total of 88 markers generated specific amplicons, whereas the other eight and four markers showed no amplification and non-specific amplification, respectively (Supplementary Fig. S7). The small number of markers detecting non-specific amplification suggested less redundancy of SSR regions in the Jatropha genome. The number of alleles per locus ranged from one to four with a mean value of 1.31. Markers showed no polymorphisms; those detecting a single allele were most frequent. PIC values ranged from 0 to 0.45 with a mean value of 0.06 (Supplementary Fig. S8). The large number of markers detecting no polymorphisms and the low mean value of the PIC indicated that genetic diversity in Jatropha lines is generally narrow. An UPGMA genetic tree of the 12 lines of J. curcas illustrated that the three lines derived from meso-America regions (Guatemala1, Guatemala2, and Mexico2b) are genetically distinct from the other lines derived from Asia and Africa, whereas no significant difference was observed between the Asian and African lines (Supplementary Fig. S9).
4. Databases
Information about the genomic sequences (contigs and singlets) and BAC clone sequences is available through international databases (DDBJ/GenBank/EMBL) under accession numbers BABX01000001–BABX01150417 (150 417 entries) and AP011961–AP011977 (17 entries), respectively. Single reads of cDNA by GS FLX sequencer derived from leaf and callus tissue are available through DDBJ Sequence Read Archive under accession numbers DRA000303 and DRA000304, respectively. Paired-end reads of genome by GAIIx sequencer with 36 bp long, and 50 and 31 bp long are available through DDBJ Sequence Read Archive under accession numbers DRA000305 and DRA000306, respectively. An online database that provides the nucleotide sequences and the predicted genes is available at http://www.kazusa.or.jp/jatropha/.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
We thank the Sumitomo Electric Industries, Ltd. for their philanthropic donation of funds to aid the Jatropha genome project celebrating the 110th anniversary of their foundation. This work was also supported by the Kazusa DNA Research Institute Foundation.
Acknowledgements
Special thanks are extended to Profs. Kazuyoshi Itoh and Yasuo Kanematsu of the Graduate School of Engineering, Osaka University, for their guidance and support during the genome project.
References
- 1.Openshaw K. A review of J. curcas: an oil plant of unfulfilled promise. Biomass Bioenergy. 2000;19:1–15. [Google Scholar]
- 2.Wouter H.M., Wouter M.J.A., Bart M. Nature Precedings; 2009. Use of inadequate data and methodological errors lead to a dramatic overestimation of the water footprint of Jatropha curcas. hdl:10101/npre.2009.3410.1 http://precedings.nature.com/documents/3410/version/1 . [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fairless D. Biofuel: the little shrub that could—maybe. Nature. 2007;449:652–655. doi: 10.1038/449652a. [DOI] [PubMed] [Google Scholar]
- 4.Biello D. Green fuels for jets. Sci. Am. 2009;19:68–69. [Google Scholar]
- 5.Carvalhoa C.R., Clarindoa W.R., Praça M.M., Araújoa F.S., Carels N. Genome size, base composition and karyotype of Jatropha curcas L., an important biofuel plant. Plant Sci. 2008;174:613–617. [Google Scholar]
- 6.Costa G.G., Cardoso K.C., Del Bem L.E., et al. Transcriptome analysis of the oil-rich seed of the bioenergy crop Jatropha curcas L. BMC Genomics. 2010;11:462. doi: 10.1186/1471-2164-11-462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sato S., Nakamura Y., Kaneko T., et al. Genome structure of the legume, Lotus japonicus. DNA Res. 2008;15:227–239. doi: 10.1093/dnares/dsn008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.White J.R., Roberts M., Yorke J.A., Pop M. Figaro: a novel statistical method for vector sequence removal. Bioinformatics. 2008;24:462–467. doi: 10.1093/bioinformatics/btm632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang X., Yang S.P., Chinwalla A.T., et al. Application of a superword array in genome assembly. Nucleic Acids Res. 2006;34:201–205. doi: 10.1093/nar/gkj419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Quinlan A.R., Stewart D.A., Strömberg M.P., Marth G.T. Pyrobayes: an improved base caller for SNP discovery in pyrosequences. Nat. Methods. 2008;5:179–181. doi: 10.1038/nmeth.1172. [DOI] [PubMed] [Google Scholar]
- 11.Gomez-Alvarez V., Teal T.K., Schmidt T.M. Systematic artifacts in metagenomes from complex microbial communities. ISME J. 2009;3:1314–1317. doi: 10.1038/ismej.2009.72. [DOI] [PubMed] [Google Scholar]
- 12.Chevreux B., Wetter T., Suhai S. Genome sequence assembly using trace signals and additional sequence information; Computer Science and Biology: Proceedings of the German Conference on Bioinformatics (GCB); 1999. pp. 45–56. 99. [Google Scholar]
- 13.Unseld M., Marienfeld J.R., Brandt P., Brennicke A. The mitochondrial genome of Arabidopsis thaliana contains 57 genes in 366,924 nucleotides. Nat. Genet. 1997;15:57–61. doi: 10.1038/ng0197-57. [DOI] [PubMed] [Google Scholar]
- 14.Zhang Z., Schwartz S., Wagner L., Miller W.A. Greedy algorithm for aligning DNA sequences. J. Comput. Biol. 2000;7:203–214. doi: 10.1089/10665270050081478. [DOI] [PubMed] [Google Scholar]
- 15.Zerbino D.R., Birney E. Velvet: algorithms for de novo short read assembly using de Bruijn graphs. Genome Res. 2008;18:821–829. doi: 10.1101/gr.074492.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kent W.J. BLAT—the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lukashin A., Borodovsky M. GeneMark.hmm: new solutions for gene finding. Nucleic Acids Res. 1998;26:1107–1115. doi: 10.1093/nar/26.4.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burge C., Karlin S. Prediction of complete gene structures in human genomic DNA. J. Mol. Biol. 1997;268:78–94. doi: 10.1006/jmbi.1997.0951. [DOI] [PubMed] [Google Scholar]
- 19.Hebsgaard S.M., Korning P.G., Tolstrup N., Engelbrecht J., Rouzé P., Brunak S. Splice site prediction in Arabidopsis thaliana pre-mRNA by combining local and global sequence information. Nucleic Acids Res. 1996;24:3439–3452. doi: 10.1093/nar/24.17.3439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Brendel V., Kleffe J. Prediction of locally optimal splice sites in plant pre-mRNA with applications to gene identification in Arabidopsis thaliana genomic DNA. Nucleic Acids Res. 1998;26:4748–4757. doi: 10.1093/nar/26.20.4748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Altschul S.F., Madden T.L., Schäffer A.A. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang X., Zhang J. Methods for comparing a DNA sequence with a protein sequence. Comput. Appl. Biosci. 1996;12:497–506. doi: 10.1093/bioinformatics/12.6.497. [DOI] [PubMed] [Google Scholar]
- 23.Ashburner M., Ball C.A., Blake J.A., et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hunter S., Apweiler R., Attwood T.K., et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ogata H., Goto S., Sato K., Fujibuchi W., Bono H., Kanehisa M. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;122:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saitou N., Nei M. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987;4:406–425. doi: 10.1093/oxfordjournals.molbev.a040454. [DOI] [PubMed] [Google Scholar]
- 28.Tamura K., Dudley J., Nei M., Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 29.Temnykh S., DeClerck G., Lukashova A., Lipovich L., Cartinhour S., McCouch S. Computational and experimental analysis of microsatellites in rice (Oryza sativa L.): frequency, length variation, transposon associations, and genetic marker potential. Genome Res. 2001;11:1441–1452. doi: 10.1101/gr.184001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steve R., Helen J.S. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 31.Bao Z., Eddy S.R. Automated de novo identification of repeat sequence families in sequenced genomes. Genome Res. 2002;12:1269–1276. doi: 10.1101/gr.88502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang B.B., Brendel V. The ASRG database: identification and survey of Arabidopsis thaliana genes involved in pre-mRNA splicing. Genome Biol. 2004;5:R102. doi: 10.1186/gb-2004-5-12-r102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bairoch A., Apweiler R. The SWISS-PROT protein sequence data bank and its new supplement TREMBL. Nucleic Acids Res. 1996;24:21–25. doi: 10.1093/nar/24.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chan A.P., Crabtree J., Zhao Q., et al. Draft genome sequence of the oilseed species Ricinus communis. Nat. Biotechnol. 2010;28:951–956. doi: 10.1038/nbt.1674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Carbon S., Ireland A., Mungall C.J., Shu S., Marshall B., Lewis S. AmiGO: online access to ontology and annotation data. Bioinformatics. 2009;15:288–289. doi: 10.1093/bioinformatics/btn615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tong L., Shu-Ming P., Wu-Yuan D., et al. Characterization of a new stearoyl-acyl carrier protein desaturase gene from Jatropha curcas. Biotechnol. Lett. 2006;28:657–662. doi: 10.1007/s10529-006-0034-3. [DOI] [PubMed] [Google Scholar]
- 37.Ye J., Qu J., Bui H.T.N., Chua N.H. Rapid analysis of Jatropha curcas gene functions by virus-induced gene silencing. Plant Biotechnol. J. 2009;7:964–976. doi: 10.1111/j.1467-7652.2009.00457.x. [DOI] [PubMed] [Google Scholar]
- 38.Saha S., Enugutti B., Rajakumari S., Rajasekharan R. Cytosolic triacylglycerol biosynthetic pathway in oilseeds. Molecular cloning and expression of peanut cytosolic diacylglycerol acyltransferase. Plant Physiol. 2006;141:1533–1543. doi: 10.1104/pp.106.082198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Haas W., Sterk H., Mittelbach M. Novel 12-deoxy-16-hydroxy phorbol diesters isolated from the seed oil of J. curcas. J. Nat. Prod. 2002;65:1434–1440. doi: 10.1021/np020060d. [DOI] [PubMed] [Google Scholar]
- 40.Lin J., Jin Y.J., Zhou X., Wang J.Y. Molecular cloning and functional analysis of the gene encoding geranylgeranyl diphosphate synthase from J. curcas. Afr. J. Biotechnol. 2010;9:3342–3351. [Google Scholar]
- 41.Aubourg S., Lecharny A., Bohlmann J. Genomic analysis of the terpenoid synthase (AtTPS) gene family of Arabidopsis thaliana. Mol. Genet. Genomics. 2002;267:730–745. doi: 10.1007/s00438-002-0709-y. [DOI] [PubMed] [Google Scholar]
- 42.Stirpe F., Pession-Brizzi A., Lorenzoni E., Strocchi P., Montanaro L., Sperti S. Studies on the proteins from the seeds of Croton tiglium and of Jatropha curcas. Toxic properties and inhibition of protein synthesis in vitro. Biochem. J. 1976;156:1–6. doi: 10.1042/bj1560001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Luo M.J., Yang X.Y., Liu W.X., et al. Expression, purification and anti-tumor activity of curcin. Acta Biochim. Biophys. Sin. 2006;38:663–668. doi: 10.1111/j.1745-7270.2006.00208.x. [DOI] [PubMed] [Google Scholar]
- 44.Lin J., Yan F., Tang L., Chen F. Antitumor effects of curcin from seeds of Jatropha curcas. Acta Pharmacol. Sin. 2003;24:241–246. [PubMed] [Google Scholar]
- 45.Qin X., Zheng X., Shao C., et al. Stress-induced curcin-L promoter in leaves of Jatropha curcas L. and characterization in transgenic tobacco. Planta. 2009;230:387–395. doi: 10.1007/s00425-009-0956-9. [DOI] [PubMed] [Google Scholar]
- 46.Qin W., Ming-Xing H., Ying X., Xin-Shen Z., Fang C. Expression of a ribosome inactivating protein (curcin 2) in Jatropha curcas is induced by stress. J. Biosci. 2005;30:351–357. doi: 10.1007/BF02703672. [DOI] [PubMed] [Google Scholar]
- 47.Huang M.-X., Hou P., Wei Q., Xu Y., Chen F. A ribosome-inactivating protein (curcin 2) induced from Jatropha curcas can reduce viral and fungal infection in transgenic tobacco. Plant Growth Regul. 2008;54:115–123. [Google Scholar]
- 48.Eitas T.K., Dangl J.L. NB-LRR proteins: pairs, pieces, perception, partners, and pathways. Curr. Opin. Plant Biol. 2010;13:472–477. doi: 10.1016/j.pbi.2010.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alvarez-Buylla E.R., Pelaz S., Liljegren S.J., et al. An ancestral MADS-box duplication occurred before the divergence of plants and animals. Proc. Natl Acad. Sci. USA. 2000;97:5328–5333. doi: 10.1073/pnas.97.10.5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rijpkema A.S., Gerats T., Vandenbussche M. Evolutionary complexity of MADS complexes. Curr. Opin. Plant Biol. 2007;10:32–38. doi: 10.1016/j.pbi.2006.11.010. [DOI] [PubMed] [Google Scholar]
- 51.Lee J.H., Yoo S.J., Park S.H., Hwang I., Lee J.S., Ahn J.H. Role of SVP in the control of flowering time by ambient temperature in Arabidopsis. Genes Dev. 2007;21:397–402. doi: 10.1101/gad.1518407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hartmann U., Höhmann S., Nettesheim K., Wisman E., Saedler H., Huijser P. Molecular cloning of SVP: a negative regulator of the floral transition in Arabidopsis. Plant J. 2000;21:351–360. doi: 10.1046/j.1365-313x.2000.00682.x. [DOI] [PubMed] [Google Scholar]
- 53.Lee S., Choi S.C., An G. Rice SVP-group MADS-box proteins, OsMADS22 and OsMADS55, are negative regulators of brassinosteroid responses. Plant J. 2008;54:93–105. doi: 10.1111/j.1365-313X.2008.03406.x. [DOI] [PubMed] [Google Scholar]
- 54.Leseberg C.H., Li A., Kang H., Duvall M., Mao L. Genome-wide analysis of the MADS-box gene family in Populus trichocarpa. Gene. 2006;378:84–94. doi: 10.1016/j.gene.2006.05.022. [DOI] [PubMed] [Google Scholar]
- 55.Dehgan B., Webster G. University of California Press, Berkeley, CA, USA; 1992. Morphology and infrageneric relationships of the genus J. curcas. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.