Abstract
Glutathione S-transferases (GSTs) exist in various eukaryotes and function in detoxification of xenobiotics and in response to abiotic and biotic stresses. We have carried out a genome-wide survey of this gene family in 10 plant genomes. Our data show that tandem duplication has been regarded as the major expansion mechanism and both monocot and dicot plants may have practiced different expansion and evolutionary history. Non-synonymous substitutions per site (Ka) and synonymous substitutions per site (Ks) analyses showed that N- and C-terminal functional domains of GSTs (GST_N and GST_C) seem to have evolved under a strong purifying selection (Ka/Ks < 1) under different selective pressures. Differential evolutionary rates between GST_N and GST_C and high degree of expression divergence have been regarded as the major drivers for the retention of duplicated genes and the adaptability to various stresses. Expression profiling also indicated that the gene family plays a role not only in stress-related biological processes but also in the sugar-signalling pathway. Our survey provides additional annotation of the plant GST gene family and advance the understanding of plant GSTs in lineage-specific expansion and species diversification.
Keywords: abiotic stress, biotic stress, comparative genomics, functional divergence, glutathione transferase
1. Introduction
Glutathione (GSH) is the tripeptide γ-glutamyl-cysteinyl-glycine and plays a central role in the processes of detoxification and redox buffering.1 Glutathione S-transferases (GSTs, EC. 2.5.1.18) catalyze the conjugation of GSH to an electrophilic substrate.2 Plant GSTs have been actively investigated during last decades.2–8 Currently, large numbers of GST genes have been identified or annotated from at least 17 plant species.7–9 In Arabdopsis, the complete identification in a genome-wide level revealed at least 53 GST genes.5,10,11 In rice, 59 GST genes have been identified.12 However, their works were carried out by BLAST searches against the rice expression sequence tag (EST) database; thus, some members may escape from their collection. Recently, 81 GST genes have been genome-widely identified in Populus trichocarpa and they exhibit extensive functional diversification.13 However, no other data have been reported on the genome-wide identification of the GST family, although at least 20 plant genomes have been completely sequenced (http://www.genomesonline.org/gold.cgi).
Since GST functions have been closely linked to stress responses, expression analyses were carried out under a wide variety of stress conditions. Evidence showed that the transcript of plant GST genes was regulated by various abiotic and biotic stresses as well as hormones including xenobiotic-type stresses, such as herbicide application,2 chilling,14 hypoxic stress,15 dehydration,16,17 wounding,18 pathogen attack,19 ethylene,20 auxin,21 2,4,6-trinitrotoluene,22 hydrogen peroxide (H2O2) and the defence signal salicylic acid (SA).23 However, whole family-based expression analyses were carried out only in Arabidopsis,11 P. trichocarpa13 and rice,24 the transcript profiling of all family members is not available for the other plants.
Although many GST genes have been isolated or annotated, only a small number of them have been functionally characterized. Reports showed that plant GSTs might play important roles in herbicide resistance2 and detoxification.25 Overexpression of GSTs in plants improved herbicide and stress tolerance,26–32 and some of GST genes have been patented.8 On the other hand, efforts have been put on the understanding of the role of GSTs in endogenous plant developmental processes. For example, studies revealed the role of GSTs in the vacuolar sequestration of anthocyanins in maize, petunia and Arabidopsis.33–36 Evidence also showed that GSTs might function as binding proteins by binding to various hormones including auxin37–39 and cytokinin40 as well as porphyrin compounds41 to regulate their activities.
Generally, although plant GSTs have been discovered for more than 30 years,5 limited data are available on the genome-wide identification and expression analysis as well as their functional divergence of the GST family. Thus, we do not know the family size in a genome and its member that are involved in biotic and abiotic stress-related biological processes. On the other hand, little is know about how these genes have been evolved or expanded with such functions. In this report, we first genome-widely identified and characterized all GST genes encoded by the sorghum and other nine plant genomes, especially focusing on the Tau and Phi classes, which are the top two subfamilies in the plant kingdom. We then evaluated their expansion and evolutionary mechanisms by investigating their duplication and/or transposition history. Subsequently, we examined their transcript profiling by the full-length cDNA, EST, microarray and massively parallel signature sequencing (MPSS42) data sets as well as by RT–PCR and quantitative real-time RT–PCR (qRT–PCR). Finally, we investigated their expression divergence under various stresses to further annotate their biological functions and retention mechanisms. Our data show that monocot and dicot GST genes exhibit differences in their evolutionary history and they have been involved in lineage-specific expansion and species diversification. Our data further confirmed their functions in biotic and abiotic stress-related developmental processes and also demonstrated that plant GSTs may be involved in the sugar-related signalling pathway.
2. Materials and methods
2.1. Plant materials and treatments
Grain sorghum (Sorghum bicolor) L. Moench cultivar BT × 623 was used for all experiments in this study. Seeds were imbibed in water. After germination, they were planted in greenhouse and were grown under natural light and temperature conditions. The 2-week-old seedlings were used for all treatments. For the drought treatment, seedlings were treated with 30% polyethylene glycol (PEG) and whole plants were collected in various time intervals (0, 0.5 and 2 h), and then frozen in liquid nitrogen for RNA preparation. For salinity and cold treatments, seedlings were subjected to the 250 mM NaCl solution and 4°C conditions, respectively. Samples for RNA extractions were collected in 0, 2 and 8 h time intervals. For glucose and sucrose treatments, seedlings were subjected to 5% of glucose and sucrose solutions, respectively, as suggested by Kojima et al.43 Samples were then collected in 0, 2 and 6 h intervals, respectively.
2.2. Expression analysis of sorghum Tau GST subfamily members by RT–PCRs and qRT–PCRs
Expression of sorghum Tau GST members was evaluated using the sorghum EST collections from several databases (http://www.ncbi.nlm.nih.gov/dbEST/index.html, http://www.phytozome.net/sorghum and http://compbio.dfci.harvard.edu/cgi-bin/tgi/est_ann.pl?gudb=Sorghum). Since low percentages of Tau members have been detected with EST expression evidence, expression analysis was carried out by normal and qRT–PCR. All gene-specific primers used for both normal and qRT–PCR analyses in this study were designed by Applied Biosystems Primer Express® software. Supplementary Table S1 lists all the primer sequences used in this study.
QIAGEN RNeasy Mini Kit was used for total RNA isolation. The first-strand cDNA was synthesized using Invitrogen kit. RT–PCRs were performed in 20 μl of reaction mixtures with 20 ng of first-strand cDNA, 200 μM of each dNTPs, 2.5 mM MgCl2, 0.5 μM each of primers, 1 U of Taq-DNA polymerase in 1× PCR buffer. These reactions were carried out using PTC-100 thermo-cyclers. The temperature profiling for PCR is as follows: 94°C for 2 min followed by 35 cycles at 94°C for 10 s, 59°C for 10 s and 72°C for 25 s followed by a 2-min extension step at 72°C. PCR products (10 μl) were visualized in 2% agarose gel, and all pictures were taken in BIORAD UV-Gel documentation system using Quantity one 1-D Analysis software. The qRT–PCR analyses were carried out according to our previous description.44
2.3. Expression divergence of rice, Arabidopsis and soybean Tau GST subfamily members
Rice Tau ESTs were collected from the Michigan State University (MSU) rice genome annotation database (http://rice.plantbiology.msu.edu/) and the NCBI EST database. Arabidopsis and soybean Tau ESTs were obtained from The Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org/) and the Soybase (http://soybase.org/index.php), respectively. The rice MPSS database (http://mpss.udel.eud/rice/)42 was used to evaluate differentially regulated genes under abiotic (drought, high salinity and cold) and biotic (Magnaporthe grisea, Mg, and Xanthomonas oryzae pv. oryzae, Xoo) stresses according to our previous description.45 For Arabidopsis, the expression data under abiotic (drought, high salinity and cold) and biotic (Erysiphe orontii and Botrytis cinerea, Bcin) stresses were downloaded, and differentially expressed Tau genes were identified according to the description.46
To evaluate the expression divergence of tandemly or segmentally duplicated genes, the Pearson's correlation coefficient (r) of their expression was computed for each pair according to the method by Lin et al.47 The cut-off r-value, below which duplicated genes can be considered divergent in their expression, might be determined by calculating r between the expression profiles of 10 000 pairs of randomly selected genes.47,48 Based on the method, we have calculated the cut-off r-value for the Arabidopsis expression data set in this study as 0.53. Thus, we utilized r < 0.53 as an indicator of divergent expression in Arabidopsis. We have used r-value = 0.59 calculated by Lin et al.47 as a cut-off in rice since we employed the same data set (MPSS) for our expression analysis. For sorghum, due to the lack of enough expression data for calculating the cut-off r-value, r < 0.5 was used as an indicator since r-values from 0.5 to 0.59 have been used for criteria to determine the diverged expression.47–49
2.4. Genome-wide identification of plant genes encoding GST proteins
The Pfam database (http://pfam.sanger.ac.uk/)50 was used to locate the GST_N (PF02798) domains in 53 key GST proteins from a variety of organisms. Similarly, the GST_C domains (PF00043) were retrieved from 58 key GST proteins from the same database. These key domain amino acid sequences were aligned by ClustalX 2.051 and were then used to generate the hidden Markov model (HMM) profiles for HMM searches with E-value cut-off of 1.0 against annotated protein databases from 10 plant organisms. These species include four monocots (Brachypodium distachyon, Oryza sativa, S. bicolor and Zea mays) and six dicots (Arabidopsis lyrata, Arabidopsis thaliana, Carica papaya, Glycine max, Medicago trunculata and P. trichocarpa). For rice, the O. sativa spp. japonica cv. Nipponbare release 6.1 pseudomolecules in the MSU Rice Annotation was used for the search. For A. thaliana, the latest version (TAIR9) of Arabidopsis genome annotation was employed (http://www.arabidopsis.org). For the remaining eight species, the latest versions were from the phytozome database (http://www.phytozome.net/). The key domain amino acid sequences were also used as queries for BLASTP/TBLASTN searches for possible homologues encoded by the 10 plant organisms with an E-value cut-off of 0.01 to confirm the HMM searches.
The presence of the domain GST_N or GST_C in the putative GST members detected by the HMM or BLAST searches was confirmed by searching the Pfam database with E-value = 0.01 as the cut-off level. The SMART database (http://smart.embl-heidelberg.de/)52 was also employed to detect conserved domains with default parameters. Proteins confirmed by domain searches were regarded as putative GST_N or GST_C domain-containing proteins (referred to GST_N or GST_C domain-containing proteins for convenience); otherwise, they were excluded from our data set.
2.5. Alignment and phylogenetic analysis
GST_N/GST_C domain or full-length amino acid sequences were aligned using ClustalX 2.0 (http://www.clustal.org/)51 and manually edited in Jalview (version 2).53 The aligned sequences were used for phylogenetic analysis according to the description by Jiang and Ramachandran.54
2.6. Expansion and evolutionary analysis
Chromosomal distributions of GST genes were performed by searching and mapping physical positions of their corresponding locus numbers in their genomes. Tandemly duplicated GST genes in 10 plant genomes were identified by three criteria (i) ≤10 genes apart, (ii) belong to the same GST subfamily and (iii) within 100 kb for both A. lyrata and A. thaliana or 350 kb for the remaining species as suggested by Lehti-Shiu et al.55 Segmentally duplicated chromosome blocks have been previously identified in a genome-wide level in Arabidopsis,56 rice57,58 and soybean (http://www.phytozome.net/soybean.php). We examined the segmental duplicates by comparing the positions of GST genes with known duplicated chromosomal blocks. For the remaining genomes, we identified duplicated blocks using the flanking regions (50 kb upstream and downstream) of GST genes according to the method by Kong et al.59
To determine the contribution of transposable elements to the expansion of the GST family, the flanking genomic sequences of the 50-kb upstream and downstream of GST genes in different genomes were used for the identification of major transposon family members including mutator-like transposable element (MULE), hAT, CACTA and Helitron families as well as the identification of retrogenes according to the description.45 Retrotransposon elements were identified by executing the LTR_Finder program60 or RetrOryza.61 Non-synonymous (Ka) and synonymous (Ks) values and their ratios (Ka/Ks) were estimated and were statistically tested according to Jiang et al.45
3. Results
3.1. The sorghum and other plant genomes encode different numbers of the GST family members
To identify and characterize the sorghum GST family members in a genome-wide level, HMM and BLAST searches were carried out against the whole sorghum genome sequences (the ‘Materials and methods’ section). Our data show that the sorghum genome encodes at least 99 GST_N or GST_C domain-containing proteins (Table 1). Supplementary Table S2 lists the details of these members including their locus names, genome positions, coding sequences and amino acid sequences. Among these two domains, GST_N contains catalytically essential residues and is thought to be a key component of catalysis in GSTs, whereas GST_C may be responsible for substrate specificity.62 Therefore, these two domains were investigated either separately or jointly to better understand their evolution or function. Based on the GST_N domain sequences, the sorghum GST family can be divided into seven different classes by phylogenetic analysis (Supplementary Fig. S1). They are Tau, Phi, Theta, Zeta, Lambda, GSH-dependent dehydroascorbate reductase and tetrachlorohydroquinone dehalogenase, as reported in other studies.5,7,11,13 Among them, both Tau and Phi are the major classes, consisting of 56 (64%) and 19 (22%) members, respectively (Table 1).
Table 1.
Genome-wide identification of GST encoding genes in the 10 completely sequenced plant genomes
| Organism | Total GSTs | Domain |
Classesa |
||||
|---|---|---|---|---|---|---|---|
| GST_N | GST_C | Both | Tau (%) | Phi (%) | Both (%) | ||
| A. lyrata | 64 | 56 | 58 | 51 | 30 (54) | 13 (23) | 43 (77) |
| A. thaliana | 60 | 54 | 51 | 45 | 28 (52) | 13 (24) | 41 (76) |
| B. distachyon | 69 | 56 | 64 | 56 | 25 (45) | 21 (38) | 46 (82) |
| C. papaya | 48 | 40 | 37 | 29 | 23 (58) | 6 (15) | 29 (73) |
| G. max | 105 | 87 | 82 | 65 | 56 (64) | 11 (13) | 67 (77) |
| M. truncatula | 43 | 40 | 36 | 33 | 31 (78) | 6 (15) | 37 (93) |
| O. sativa | 84 | 77 | 75 | 68 | 50 (65) | 18 (23) | 68 (88) |
| P. trichocarpa | 81 | 81 | 81 | 81 | 56 (69) | 9 (11) | 65 (80) |
| S. bicolor | 99 | 87 | 87 | 78 | 56 (64) | 19 (22) | 75 (86) |
| Z. mays | 72 | 67 | 58 | 53 | 37 (55) | 23 (34) | 60 (90) |
aClassification was based on the GST_N domain amino acid sequences.
In addition to the sorghum GST family, we have also genome-widely identified GST members from other nine genomes as described in the ‘Materials and methods' section. Our data show that the sorghum and other plant genomes encode different numbers of the GST family members, and both Tau and Phi classes of GST members consist of at least 73% of total GSTs as shown in Table 1. Their locus names, genome positions, coding sequences and amino acid sequences were included in Supplementary Table S2. Currently, the GST members from Arabidopsis, rice and poplar have been reported12,13,63 and members from the remaining 7 plant genomes were newly identified. Among the 53 previously identified GSTs in Arabidopsis (http://www.arabidopsis.org/browse/genefamily/gst.jsp),63 two of them, AtT3g55040 and At5g02780, were not included in this study since no GST_N or GST_C domains could be detected with E-value = 0.01 as the cut-off level. However, we have identified nine more GST_N or GST_C members (Supplementary Table S3). Soranzo et al.12 have identified 59 rice GST members. However, three members OsGSTMU357, OsGSTU37 and OsGSTT2 were not included in this study since we cannot detect any GST domains in these members and we have also identified 28 more GST members (Supplementary Table S4). On the other hand, we have identified the same numbers of the poplar GST members as reported by Lan et al. (2009)13 and we have used them for this study.
Besides the full-length GST_N and/or GST_C domain-containing genes, we have also identified numbers of partial GST fragments. They encode only partial GST_N or GST_C or both of partial domains. These members contain no typical domain structure and have no expression evidence with the characters of pseudogenes. Due to the low feasibility of phylogenetic analyses by integrating these partial fragments, we removed these members from our analyses although we may underestimate the rate of gene duplication.
3.2. A significantly larger Tau and Phi subfamily of most recent common ancestor in monocots than in dicots
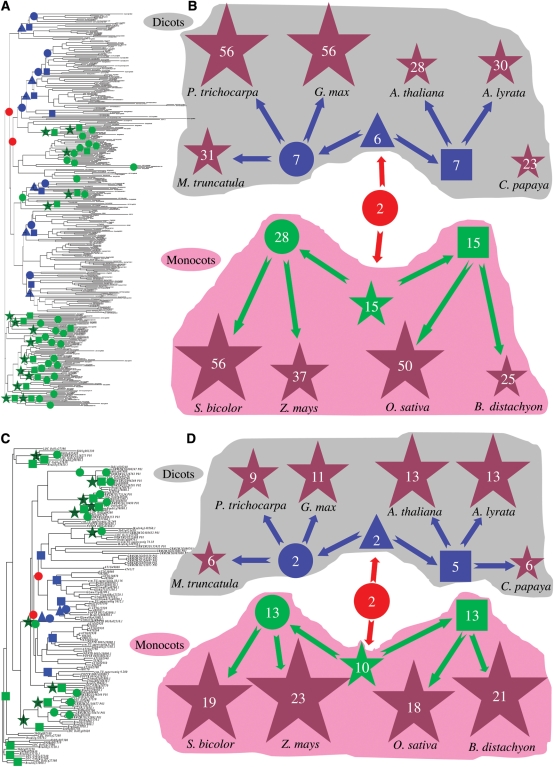
Since both Tau and Phi classes are the largest subfamilies in all analysed 10 genomes, we evaluated the patterns of expansion of these two classes by analysing the phylogenetic relationships of these GST members from different organisms (Fig. 1A–D and Supplementary Fig. S2 and S3). Most of dicot members fell into their own subclasses, separating from monocot plants. Members from each species intended to be clustered together, exhibiting lineage-specific expansion. To determine the degrees of expansion of these two classes among monocot and dicot plant lineages, we broke down the phylogeny into ancestral units according to the method.64 Due to possible gene loss and pseudogenes, the most recent common ancestor (MRCA) members may be underestimated. Thus, at least 2 Tau or Phi members would have been present in the MRCA among these 10 organisms (red circles in Fig. 1A and B for Tau and C and D for Phi). The MRCA among the 4 monocot plants possessed 15 Tau (green stars in Fig. 1A and B) and 10 Phi (green stars in Fig. 1C and D) members; thus, 7.5-fold and 5-fold expansions have occurred for Tau and Phi, respectively, in this period. Following this era, they underwent relatively lower expansion rates except for rice, resulting in the current Tau or Phi members. These results suggested that the rapid gene expansion for the 4 monocots occurred after the divergence of monocot from dicot plants. However, the MRCA among the 6 dicot plants contained only 6 Tau (blue triangles in Fig. 1A and B) and 2 Phi (blue triangles in Fig. 1C and D) members; thus, only 3-fold or no expansion has occurred for Tau and Phi, respectively, in this period. The large-scale expansion of Tau or Phi members for dicot plants occurred during their divergence from their MRCA (Eurosid I and II) (Fig. 1). These data demonstrate that both monocot and dicot plants exhibit the differences in their Tau and Phi subfamily expansion history.
Figure 1.
Phylogenetic analysis and evolutionary dynamics of the Tau and Phi classes of the GST superfamily. (A and C) Phylogenetic analyses of the Tau and Phi subfamily members, respectively, in four monocot and six dicot plants. GST_N domain amino acid sequences were employed to construct phylogenetic trees using the bootstrap method with a heuristic search of the PAUP 4.0b8 program. The results were confirmed by the Bayesian analyses. Ancestral units were defined according to Shiu et al.64 Their enlarged phylogenetic trees and their analyses are shown in Supplementary Figs S2 and S3, respectively. (B and D) Evolutionary history of the Tau and Phi subfamily members in 10 organisms, respectively. Red circles represent the MRCA Tau/Phi units among all 10 organisms, blue triangles indicate the MRCA Tau/Phi units among dicot plants and green stars show the MRCA Tau/Phi units among monocot plants. Blue circles and squares represent the MRCA Tau/Phi units in Eurosid I (M. truncatula, P. trichocarpa and G. max) and Eurosid II (A. thaliana and A. lyrata), respectively. Green circles and squares show the MRCA Tau/Phi units between S. bicolor and Z. mays as well as between O. sativa and B. distachyon, respectively. Brown stars indicate the expanded Tau/Phi members in all 10 organisms. Grey and pink shadows in (B) and (D) indicate dicot and monocot plant species and their MRCAs, respectively.
3.3. Significant contributions of tandem duplications to family size
To explore the possible mechanisms of the Tau and Phi GST subfamily expansion, we investigated the contributions of both tandem and segmental duplications to the expansion of these subfamilies in the sorghum genome. We examined the physical positions of the Tau and Phi GST members on different sorghum chromosomes (Supplementary Fig. S4). The results showed that both Tau/Phi GSTs were located on multiple chromosomes with non-random distributions. For example, chromosome 1 contained the highest density of the Tau or Phi GSTs with 30 or 7 members, respectively. Similar results were observed in rice and Arabidopsis.5,12 However, more even distributions were observed in soybean with fewer members in most of chromosomes (data not shown). Based on the physical positions of the Tau or Phi GSTs, the tandem cluster was defined as described in the Methods. Among the 56 Tau and 19 Phi GSTs in sorghum, 44 Tau (78%) and 12 Phi (63%) GSTs are found in tandem clusters, respectively (red locus name in Supplementary Fig. S4; Fig. 2), indicating tandem duplications as the main mechanism for the Tau and Phi GST expansion. On the other hand, only two members of Tau and Phi GSTs (Sb01g005990 and Sb01g013590 for Tau and Sb01g030240 and Sb01g047980 for Phi) were involved in segmental duplications (indicated by blue lines in Supplementary Fig. S4). Besides both tandem and segmental duplications, we also investigated the contributions of other expansion mechanisms including transpositions and retrotranspositions to the expansion of these subfamilies (see Methods). However, our data showed that limited contribution by both transposons and retrotransposons to the expansion was detected. Similarly, we have investigated the contributions of tandem and segmental duplication, transpositions and retrotranspositions to the expansion of Tau and Phi subfamilies in the remaining 9 plant species (Fig. 2). The results showed that the expansion of 46–82% of Tau and 33–76% Phi GSTs were found in tandem clusters (Fig. 2), suggesting tandem duplication as the major mechanism for Tau and Phi GST expansion. In addition, 2–23% of Tau and 0–72% Phi GSTs were segmentally duplicated (Fig. 2), suggesting that segmental duplication should also be regarded as the major expansion mechanism for the Phi class in some species such as in soybean and Arabidopsis.
Figure 2.
Pie diagrams showing expansion mechanisms of the Tau and Phi classes of the GST superfamilies in 10 plants. The figure indicates the contributions of tandem (red), segmental (green) duplications and both of them (pink) as well as other mechanisms (blue) to the expansion of the Tau and Phi class members in 10 genomes. In each pie diagram, the number prior to the comma indicates the total expanded members by different expansion mechanisms and its percentage is given following the comma.
Since tandem duplication is regarded as the major expansion mechanism, we re-constructed their expansion history using the largest tandem cluster with 23 members in the sorghum chromosome 1. The 23 tandem genes are in two clades (Supplementary Fig. S5A), suggesting that this cluster is the results of two ancestral units, which may be evolved from ancient tandem duplication events. One of them contains only 3 genes whereas the secondary clade contains 20 members. The data suggested that different ancestral genes duplicated themselves by differential expansion rates. On the basis of the phylogenetic tree, we deduced the hypothetical origins of 23 genes from two ancestral units by tandem duplication (Supplementary Fig. S5B). For the ancestral unit with 3 descendents, only two rounds of tandem duplications were required to generate these genes. However, for the secondary unit with 20 descendents, at least 9 rounds of tandem duplications were expected to produce such a population. After expansion, these genes were then not always inserted into the loci according to their physical orders. For example, the putative tandem pair of the Sb01g030810 gene is Sb01g030990 but not its physical neighbour Sb01g030820. On the other hand, we found that most of tandem duplications occurred by a one-gene mode, i.e. only one gene was duplicated in a one tandem duplication event. This case was also observed in our previous report on tandem duplication of the lectin gene superfamily.65 Taken together, our data showed that parental genes were not always physically linked to their descendant genes and different expansion rates were observed for different parental genes.
3.4. Different selection forces between monocot and dicot plants and between GST_N and GST_C domains in a species
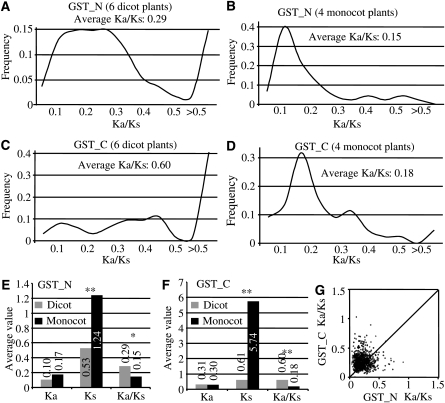
Since both monocot and dicot plants exhibited the distinct difference in their expansion history, we investigated whether they were under different selection forces. We first identified reciprocal best matches for all Tau or Phi GST members either from 4 monocot or 6 dicot plants. These identified matches were then used to calculate Ka/Ks ratios for their GST_N domain (Fig. 3A and B). Evolutionarily, the ratio of Ka to Ks can be used as an indicator of selective pressure acting on a protein-coding gene. A Ka/Ks ratio <1 indicates functional constraint with purifying selection on a gene, and a Ka/Ks ratio >1 indicates accelerated evolution with positive selection and a Ka/Ks ratio = 1 indicates neutral selection. Although the period with large scale of expansion of Tau or Phi GSTs from 6 dicot plants was later than 4 monocot plants, the average Ka/Ks for 6 dicots (0.29) is significantly higher than that for 4 monocots (0.15). Furthermore, the frequency distribution of the Ka/Ks ratios is also significantly different between dicot and monocot plants. For 6 dicots, most of the mass were centred near Ka/Ks = 0.2 or >0.5 whereas most of the mass for 4 monocots were centred near Ka/Ks = 0.12. These data suggested that GST_N in dicot plants evolved faster than that in monocot plants. Similar results were observed when GST_C domain regions were used for such analyses (Fig. 3C and D). To further analyse the reason why both dicot and monocot plants exhibited the difference in their evolutionary rates, we compared their Ka and Ks values separately. We found that their Ka values between dicot and monocot plants were similar (Fig. 3E and F). However, both dicot and moncot plants exhibited significant differences in their Ks values (Fig. 3E and F). As a result, they showed significant differences in their Ka/Ks ratios.
Figure 3.
Ka/Ks ratio analysis. Frequency distributions of Ka/Ks ratios were analysed using the best matched pairs among six dicot plants (A and C) or four monocot plants (B and D). (A) and (B) show the data analysed from the GST_N regions. (C) and (D) show the analysis from the GST_C regions. The average Ka, Ks and their ratios in monocot and dicot pairs were indicated in (E) for GST_N and in (F) for GST_C. Asterisks indicate significant differences between monocot and dicot plants at P < 0.05 (*) and P < 0.01 (**) by t-test, respectively. The Ka/Ks plots for the GST_C versus GST_N domains of the sorghum Tau members are shown in (G).
Since most of plant GST proteins consist of two functional domains including GST_N and GST_C, we were wondering if these two domains exhibited different evolutionary rates. We analysed the Ka/Ks ratios of these two domains of the sorghum Tau members separately. The ratios of GST_C plotted against that of GST_N of same proteins were shown in Fig. 3G. The results suggested that both domains were under differential selective pressures and the GST_C domain might have been subjected to more relaxed functional constraints. Similar results were observed in the remaining 9 organisms (data not shown). The recent analysis on the Populus GST superfamily also showed similar results.13
3.5. Expression abundance of the Tau subfamily members among 9 sorghum tissues
Tau class is the largest GST subfamily in plants and is a good candidate for surveying the expression divergence of duplicated members. Therefore, we further investigated the expression profiling of this class. We have detected 33 out of 56 sorghum Tau members with EST expression evidence. To examine if the remaining genes are expressed in sorghum and to explore their expression divergence, we have investigated the expression of 50 annotated Tau genes in 9 different tissues including young and mature leaves, panicles, seeds and roots as well as stems by RT–PCR analysis. The results showed that not all annotated Tau genes were expressed under normal growth conditions. Among 50 tested Tau members, 37 genes (74%) were expressed in at least one of 9 tissues (Fig. 4) and the remaining 13 genes showed no expression in all tested tissues. All these results have been confirmed by qRT–PCR (Supplementary Fig. S6). Among the 37 expressed genes, nine of them were expressed in all tested tissues. These genes were listed as follows: 01g030800, 01g030810, 01g030990, 02g022210, 02g038130, 03g045780, 03g045830, 03g045860 and 05g001525. Interestingly, 10 genes showed root-specific expression including 01g027620, 01g030830, 01g030880, 01g030890, 01g030940, 01g031010, 01g031020, 01g031040, 01g031050 and 03g031780. They were mainly expressed in either young or mature roots. On the other hand, we have detected two genes with leaf-specific expression patterns including 01g030840 and 03g025210.
Figure 4.
Transcript profiles of the sorghum Tau class of GST superfamily members in nine different sorghum tissues shown by RT–PCR analyses. Nine bands from left to right for each gene represent amplified products from young leaves, mature leaves, young roots, mature roots, young panicles, mature panicles, young seeds, mature seeds and stems, respectively. The amplification of the gene SbUBQ5 with the locus name Sb09g004630 was used as control in this study as this gene showed the similar expression level among different tissues or under various stresses in this study.
3.6. Regulated expression of the Tau subfamily members in sorghum under various abiotic stresses and sugar treatments
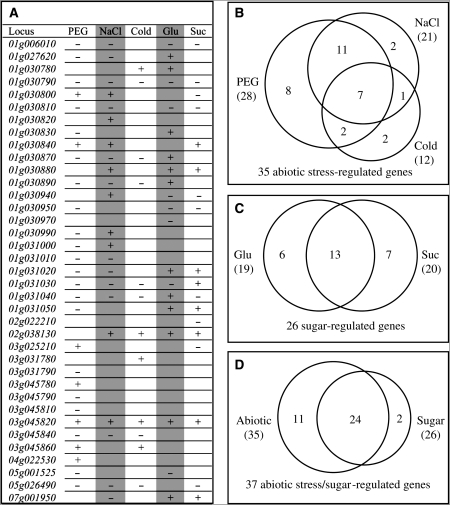
To investigate if some members are expressed only under stressed conditions and to explore their roles and functional divergence, RT–PCRs were carried out to detect their expression abundance under various abiotic stress conditions (cold, PEG or high salinity) and sugar (glucose and sucrose) treatments. The results indicated that 37 Tau members showed expression under one or more stressed conditions (Supplementary Fig. S7A). All of them were also expressed under normal growth conditions (Fig. 4). The results were confirmed by qRT–PCR (data not shown). Furthermore, we have detected additional 6 genes with significant expression signals under various stresses (Supplementary Fig. S7B). Most of them showed low expression level, which may explain why their signals could not be detected by normal RT–PCR. Thus, we have detected total of 43 GST Tau members with detectable expression signals under normal or stressed conditions.
By comparing the RT–PCR with qRT–PCR results and by statistic analysis, 37 genes were detected with significant differences in their expression level under one or more stress conditions or treatments. These genes and their responses to stresses and treatments were shown in Fig. 5A. Total of 35 Tau genes were significantly regulated by abiotic stresses including cold, PEG and high salinity (Fig. 5B). Among them, 12 genes were regulated by only one of these stresses (8 by PEG and 2 by salinity or cold). Another set of 14 genes were regulated by two of these three stresses and the remaining 7 genes were regulated by all three stresses. Besides abiotic stresses, sugar treatment was also shown to play important roles in regulating Tau GST gene expression. Totally, 26 Tau GST members were detected with significant differences in their expression level under both glucose and sucrose treatments (Fig. 5C). Among them, 6 genes were regulated by only glucose treatment and 7 genes were by sucrose. The remaining 13 genes were regulated by both treatments.
Figure 5.
A summary of the expression analyses of the sorghum Tau class of GST superfamily members under various treatments. (A) A list of expression patterns of 37 Tau members under five different treatments. The results were from RT–PCR and two biologically replicated qRT–PCR analyses. The ‘+’ indicates that the corresponding gene was significantly up-regulated under a stress treatment by t-test. The ‘−’ indicates down-regulation by t-test. (B) to (D) Venn diagrams showing the classification of genes regulated by various abiotic stresses and sugar treatments based on the result from (A).
Among 37 abiotic stress or sugar-responsive Tau members, 11 genes were regulated by only abiotic and 2 genes were by sugar; the remaining 24 genes were regulated by both abiotic and sugar treatments (Fig. 5D). These data suggested the possible interaction between abiotic stresses and sugar signalling.
3.7. Expression divergence among tandemly and fragmentally duplicated genes in sorghum, rice and Arabidopsis
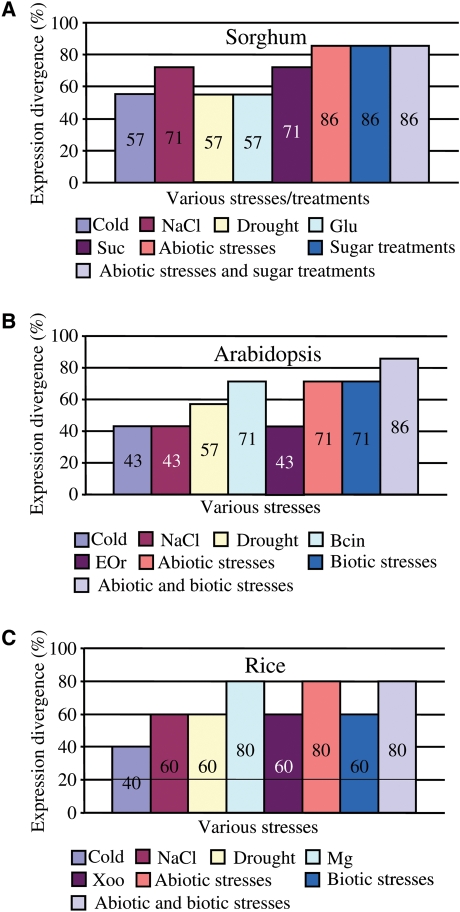
To understand the functional divergence of duplicated Tau genes, we compared the expression profiles of these genes under various stresses or different sugar treatments in sorghum. Totally, 7 tandem arrays and 1 pair of segmentally duplicated genes were submitted for such analyses. If r < 0.5, 0.59, and 0.53 (see Methods) between a tandem array or a segmental pair in sorghum, rice and Arabidopsis, respectively, the tandem array or segmental pair was regarded as a divergent array or pair in their expression. Based on the criteria, no expression divergence has been observed between segmentally duplicated gene pair 01g005990 and 01g013590. Among 7 tandem arrays, 57–71% of them exhibited significant divergence in their expression patterns in response to cold, NaCl, PEG, glucose or sucrose treatments (Fig. 6A). Statistically, 86% of arrays showed significant divergence under abiotic stresses, sugar treatments or both of them (Fig. 6A). More detail analysis by comparing the expression data with phylogenetic relationship showed that expression divergence was observed even in closely related genes.
Figure 6.
Expression divergence among tandemly and fragmentally duplicated Tau genes in sorghum, rice and Arabidopsis. (A) The effect of tandem and segmental duplications on gene expression divergence of the Tau subfamily members under abiotic stresses and sugar treatments. (B and C) Gene expression divergence of tandemly/fragmentally duplicated Tau members under abiotic and biotic stresses in rice and Arabidopsis, respectively.
Similarly, we have also investigated the expression divergence of duplicated Tau genes in Arabidopsis and rice, respectively (Fig. 6B and C). For Arabidopsis, we have analysed total of 3 pairs of segmentally duplicated pairs (At1g10360 and At1g59670, At1g17190 and At1g78340, At1g27130 and At1g69920), two of them (67%) showed expression divergence in response to either abiotic or biotic stresses. Among 7 tandem arrays, 43–71% of them were differentially expressed under cold, NaCl, PEG, Bcin or EQr treatments (Fig. 6B). As a result, 71% of them showed significant divergence under abiotic or biotic stresses and 86% of them exhibited expression divergence under both abiotic and biotic stresses (Fig. 6B). For rice, two pairs of segmentally duplicated Tau genes (LOC_Os03g57200 and LOC_Os07g05800, LOC_Os11g03210 and LOC_Os12g02960) have been detected. The former pair exhibited divergence in their transcript abundance among various tissues and no expression divergence has been detected in the latter pair. On the other hand, among 5 tandemly duplicated Tau gene arrays, 40–80% of them were divergent in their expression under cold, NaCl, PEG, Mg or Xoo treatments (Fig. 6C). Thus, 60–80% of them exhibited expression divergence under either abiotic/biotic or both of them (Fig. 6C).
4. Discussion
4.1. Evolutionary origins of GST domains and their combinations
In this study, we have genome-widely identified more than 700 GST_N or GST_C domain-containing GSTs from 10 higher plants. Besides these, our searches and reports show that GSTs are ubiquitous not only in higher plants but also in other eukaryotes and prokaryotes.62,66 Thus, GSTs are evolutionarily ancient proteins and one may be interested in the origin of this family. Reports suggested Theta, Zeta and Omega GSTs as the most ancestral classes.66,67 Our phylogenetic analysis also supported both Theta and Zeta as the ancestral classes in plants (Supplementary Fig. S1). On the other hand, GSTs were thought to have evolved from a thioredoxin-like ancestor in response to the development of oxidative stress68,69 and glutaredoxins are the suggested ancestors of the GST_N domain.70 In this study, we indeed detected several GST_N domains, for example, in the sorghum locus Sb02g003090, with very high sequence similarity to the glutaredoxin domain, supporting the putative origin of GSTs from glutaredoxins.
Among these identified hundreds of GSTs, both GST_N and GST_C domains are usually encoded in a single gene, indicating that most of GSTs are homodimers in plants. The fact also suggests the co-evolution of both GST_N and GST_C domains. However, we have also identified some GST genes encoding only GST_N or GST_C domain (Table 1). The fact may imply the loss of GST_N or GST_C domain during long evolution. One of such examples was shown in Supplementary Fig. S8A. The figure shows a tandem cluster consisting of 4 rice GSTs and their domain organizations. After duplication, only the GST_C domain was detected in one of duplicates LOC_Os10g38150 whereas both domains could be detected in the remaining tandemly duplicated genes.
Besides the detection of putative domain loss events, we are also interested in the detection of domain combinations, which may generate new proteins and thereby functional divergences.71,72 Our data showed that most of GSTs contained only GST-N/GST_C or both of them, suggesting the limited domain combination in this family. However, we have detected some exceptions. For example, both members of the Theta class Sb04g007760 and Sb10g022570 contained the Elongation factor 1 gamma domain (EF1G) except for both GST_N and GST_C domains (Supplementary Fig. S8B). These cases were also detected in Arabidopsis (Supplementary Fig. S8B) and other plants (data not shown). In Arabidopsis, we have also detected the integration of other domains such as EF-1 guanine nucleotide exchange domain (EF1_GNE) and Myb transcription factor domain (Myb_DNA-binding) (Supplementary Fig. S8B). The integration of the transcription factor domain resulted in the localization of this protein to the nucleus,73 suggesting the functional divergence by domain combination.
4.2. Differential evolutionary history of the Tau and Phi subfamilies in monocot and dicot plants
In eukaryotes, most of genes with structural and regulatory functions are members of gene families. They are descendants from gene duplication, which plays a major role in plant evolution.74 These duplicates may be lost or be survival by either retaining their original functions, subfunctionalization or neofunctionalization.75 However, limited information is available on the patterns of functional diversification governing the evolution of most classes of gene families in the plant kingdom.13
Sorghum has been regarded as a biofuel crop of growing importance for ethanol production. Now both methylation filtration-based genome sequencing and 8-time-assembly draft sequencing have been finished and their data have been freely released.76,77 Thus, it is now becoming more feasible and imperative to further characterize its genes and their families for better understanding their evolutionary mechanisms. However, comparing with other plants, limited data has been reported on the genome-wide analysis of a gene family in sorghum. We have constructed a hypothetical evolutionary history of the Tau and Phi classes of the GST family and found numbers of pseudogenes with partial domain structures, indicating that some duplicated genes failed to persist similar to other gene families.75 On the other hand, we found that the era for a large-scale expansion was different between dicot and monocot plants. Our data from 6 dicot and 4 monocot plant genomes showed that different dicot plants exhibited similar evolutionary patterns, so did monocot plants. The MRCA of all analysed dicot plants had a small family of the Tau and Phi GST subfamilies and the MRCA of all monocot plants had evolved into bigger sizes of these subfamilies (Fig. 1). The data suggested that the MRCA of monocot plants duplicated Tau and Phi GST members faster than the MRCA of dicot plants. However, during the period from the MRCA of monocot or dicot plants to current species, dicot plants evolved faster than monocot plants with more rapid expansion and higher Ka/Ks ratios (Figs. 1 and 3). Our data also show that sorghum plants exhibit high percentages of regulated expression patterns among different tissues and under various abiotic stresses (Figs. 4 and 5) and both monocot and dicot plants have also been detected with high expression divergence under these stresses (Fig. 6). Therefore, it is reasonable to suggest that differential evolutionary history between monocot and dicot plants may be due to the changed adaptability to various environmental conditions during the divergence of monocot plants from dicot plants.
4.3. Lineage-specific expansion and species diversification
We have detected two large-scale expansion events of the Tau or Phi subfamily by investigating the expansion history of these classes in 6 dicot and 4 monocot plants. The MRCA of monocot plants experienced the first large-scale expansion with 7.5-fold increase for Tau and 5-fold for Phi while dicot plants required only 3-fold more members for Tau or no expansion for Phi GSTs (Fig. 1). The second large-scale expansion occurred mainly in dicot plants with the maximum 8-fold expansion during the species diversification of some dicot plants from their common ancestors (Fig. 1). These results suggested that the large-scale expansions might be required for species diversification although more GST members from such expansion might be also for the environmental adaptability or biological processes. On the other hand, we have also detected some species-specific Tau or Phi members as reported by Lan et al.13 In fact, Tau or Phi classes of GSTs are plant-specific and species-specific GST members have been detected not only in plants but also in non-plant organisms.5,7,8 Thus, both evolutionary history and lineage-specific expansion suggested that GST members may have contributed to species diversification.
4.4. Expansion patterns and mechanisms of duplications
We have demonstrated that dicot and monocot plants exhibited different expansion patterns. We have also shown that tandem duplication represented the major mechanism for the subfamily expansion. At least eight classes of the GST subfamily have been identified7 and they exhibited different rates of expansion, resulting in different sizes of classes. Further investigation showed that the sizes of subfamilies were proportional to the tandem duplication rates (data not shown). The results suggested that tandem duplication could be regarded as the major driver for the expansion of not only the Tau/Phi GST subfamily but also the other subfamilies.
On the other hand, the rice genome has been reported to undergo large-scale duplication 40–50 million years ago78 and rice is likely an ancient aneuploid rather than a polyploidy57; however, we do not see evidence for the significant contribution of large-scale duplications within a relative short period to the expansion of the rice GST family. On the other hand, we also investigated the contributions of other mechanisms to the expansion of this family since different organisms have evolved into different sizes of GST families. We have identified all possible transposons (MULE, hAT, CACTA and Helitron) and retrotransposons (LTR and non-LTR) using flanking genomic sequences of the 50-kb upstream and downstream of GST members in four monocot and six dicot plants. We found very limited contribution of transposons/retrotransposons to the expansion of this family in multiple organisms. For example, only one of the rice Tau members LOC_Os10g38710 was located in a PACK-MULE member TI0007202, which was identified by Juretic et al.79 However, we cannot detect its parental gene.
4.5. Expression divergence and biological functions
At least 26 sorghum Tau members were regulated by glucose or sucrose treatments (Fig. 5C). In Arabidopsis, we have also detected five Tau genes with up-regulated expression under sucrose treatment based on the microarray analysis.80 These data suggested that the Tau subfamily members in plants might play a role in sugar signalling.
The complexity of the gene family functions was mainly reflected by the very high expression divergence under various stresses.81 Our data revealed that the Tau subfamily has evolved into a highly divergent group in their expression under various abiotic stresses and sugar treatments. These phenomena have been observed in all tested organisms including sorghum, rice and Arabidopsis (Fig. 6). Furthermore, the expression divergence was also observed among different individuals within a tandem array. For example, among 23 tandem duplicates (Supplementary Fig. S5), 20 GST genes show detectable expression and none of them exhibit the same expression pattern when their expression patterns were examined among different tissues (Fig. 4) or under different stresses/treatments (Fig. 5 and Supplementary Fig. S7). In addition, expression divergence of the Tau members was observed not only under these stresses but also under various hormones. For example, in sorghum, we have detected at least 40% of tandemly duplicated Tau genes with diverse expression patterns under various hormones including SA, methyl jasmonate or the ethylene precursor aminocyclopropane based on the microarray data.82 All these data suggested that expression divergence should be regarded as one of the major drivers to facilitate the retention of the tandemly duplicated genes in this subfamily. Additionally, these data also suggested that the Tau subfamily members should play important roles in stress regulation as well as sugar and hormone signalling.
On the other hand, functions of plant Tau and Phi members may be retained with a relatively low divergence as shown by Ka/Ks analysis (Fig. 3). We also showed that both GST_N and GST_C domains were under different selective pressures (Fig. 3). GSTs have been thought to function in detoxification of xenobiotics and in response to biotic and abiotic stresses. Thus, both more relaxed functional constraints in the GST_C domain and high degree of expression divergence should be regarded as the major mechanisms to facilitate the retention of duplicated genes and the adaptability to the diversity of potential xenobiotics and stressors.
Supplementary data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
This work was supported by Strategic Research Program from Temasek Life Sciences Laboratory in Singapore.
Acknowledgements
We thank the Joint Genome Institute, the MSU Buell Lab and the Arabidopsis Information Resource for providing annotations and sequences. We also thank the National Center for Biotechnology Information, the rice MPSS database, the RIKEN Plant Science Center, the DFCI Sorghum bicolor Gene Index, the Soybase and Texas A&M University for making the EST and/or stress expression data sets available.
References
- 1.Noctor G., Foyer C.H. Ascorbate and glutathione: keeping active oxygen under control. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1998;49:249–79. doi: 10.1146/annurev.arplant.49.1.249. doi:10.1146/annurev.arplant.49.1.249. [DOI] [PubMed] [Google Scholar]
- 2.Edwards R., Dixon D.P., Walbot V. Plant glutathione S-transferases: enzymes with multiple functions in sickness and in health. Trends Plant Sci. 2000;5:193–8. doi: 10.1016/s1360-1385(00)01601-0. doi:10.1016/S1360-1385(00)01601-0. [DOI] [PubMed] [Google Scholar]
- 3.Marrs K.A. The functions and regulation of glutathione S-transferases in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1996;47:127–58. doi: 10.1146/annurev.arplant.47.1.127. doi:10.1146/annurev.arplant.47.1.127. [DOI] [PubMed] [Google Scholar]
- 4.Droog F. Plant glutathione S-transferases, a tale of theta and Tau. J. Plant Growth Regul. 1997;16:95–107. doi:10.1007/PL00006984. [Google Scholar]
- 5.Dixon D.P., Lapthorn A., Edwards R. Plant glutathione transferases. Genome Biol. 2002;3:3004.1–10. doi: 10.1186/gb-2002-3-3-reviews3004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Moons A. Regulatory and functional interactions of plant growth regulators and plant glutathione S-transferases (GSTs) Vitam. Horm. 2005;72:155–202. doi: 10.1016/S0083-6729(05)72005-7. doi:10.1016/S0083-6729(05)72005-7. [DOI] [PubMed] [Google Scholar]
- 7.Basantani M., Srivastava A. Plant glutathione transferases: a decade falls short. Can. J. Bot. 2007;85:443–56. doi:10.1139/B07-033. [Google Scholar]
- 8.Chronopoulou E.G., Labrou N.E. Glutathione transferases: emerging multidisciplinary tools in red and green biotechnology. Recent Pat. Biotechnol. 2009;3:211–23. doi: 10.2174/187220809789389135. doi:10.2174/187220809789389135. [DOI] [PubMed] [Google Scholar]
- 9.Conn S., Curtin C., Bézier A., Franco C., Zhang W. Purification, molecular cloning, and characterization of glutathione S-transferases (GSTs) from pigmented Vitis vinifera L. cell suspension cultures as putative anthocyanin transport proteins. J. Exp. Bot. 2008;59:3621–34. doi: 10.1093/jxb/ern217. doi:10.1093/jxb/ern217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sappl P.G., Heazlewood J., Millar A.H. Untangling multi-gene families in plants by integrating proteomics into functional genomics. Phytochemistry. 2004;65:1517–30. doi: 10.1016/j.phytochem.2004.04.021. doi:10.1016/j.phytochem.2004.04.021. [DOI] [PubMed] [Google Scholar]
- 11.Sappl P.G., Carroll A.J., Clifton R., Lister R., Whelan J., Harvey Millar A., Singh K.B. The Arabidopsis glutathione transferase gene family displays complex stress regulation and co-silencing multiple genes results in altered metabolic sensitivity to oxidative stress. Plant J. 2009;58:53–68. doi: 10.1111/j.1365-313X.2008.03761.x. doi:10.1111/j.1365-313X.2008.03761.x. [DOI] [PubMed] [Google Scholar]
- 12.Soranzo N., Sari Gorla M., Mizzi L., De Toma G., Frova C. Organisation and structural evolution of the rice glutathione S-transferase gene family. Mol. Genet. Genomics. 2004;271:511–21. doi: 10.1007/s00438-004-1006-8. doi:10.1007/s00438-004-1006-8. [DOI] [PubMed] [Google Scholar]
- 13.Lan T., Yang Z.L., Yang X., Liu Y.J., Wang X.R., Zeng Q.Y. Extensive functional diversification of the Populus glutathione S-transferase supergene family. Plant Cell. 2009;21:3749–66. doi: 10.1105/tpc.109.070219. doi:10.1105/tpc.109.070219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Seppanen M.M., Cardi T., Hyokki M.B., Pehu E. Characterisation and expression of cold-induced glutathione S-transferase in freezing tolerant Solanum commersonii, sensitive S. tuberosum and their interspecific somatic hybrids. Plant Sci. 2000;153:125–33. doi: 10.1016/s0168-9452(99)00252-6. doi:10.1016/S0168-9452(99)00252-6. [DOI] [PubMed] [Google Scholar]
- 15.Moons A. Osgstu3 and osgtu4, encoding tau class glutathione S-transferases, are heavy metal- and hypoxic stress-induced and differentially salt stress-responsive in rice roots. FEBS Lett. 2003;553:427–32. doi: 10.1016/s0014-5793(03)01077-9. doi:10.1016/S0014-5793(03)01077-9. [DOI] [PubMed] [Google Scholar]
- 16.Kiyosue T., Yamaguchi-Shinozaki K., Shinozaki K. Characterization of two cDNAs (ERD11 and ERD13) for dehydration-inducible genes that encode putative glutathione S-transferases in Arabidopsis thaliana L. FEBS Lett. 1993;335:189–92. doi: 10.1016/0014-5793(93)80727-c. doi:10.1016/0014-5793(93)80727-C. [DOI] [PubMed] [Google Scholar]
- 17.Bianchi M.W., Roux C., Vartanian N. Drought regulation of GST8, encoding the Arabidopsis homologue of ParC/Nt107 glutathione transferase/peroxidase. Physiol. Plant. 2002;116:96–105. doi: 10.1034/j.1399-3054.2002.1160112.x. doi:10.1034/j.1399-3054.2002.1160112.x. [DOI] [PubMed] [Google Scholar]
- 18.Vollenweider S., Weber H., Stolz S., Chetelat A., Farmer E.E. Fatty acid ketodienes and fatty acid ketotrienes: Michael addition acceptors that accumulate in wounded and diseased Arabidopsis leaves. Plant J. 2000;24:467–76. doi: 10.1046/j.1365-313x.2000.00897.x. doi:10.1046/j.1365-313x.2000.00897.x. [DOI] [PubMed] [Google Scholar]
- 19.Mauch F., Dudler R. Differential induction of distinct glutathione-S-transferases of wheat by xenobiotics and by pathogen attack. Plant Physiol. 1993;102:1193–201. doi: 10.1104/pp.102.4.1193. doi:10.1104/pp.102.4.1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhou J., Goldsbrough P.B. An Arabidopsis gene with homology to glutathione S-transferases is regulated by ethylene. Plant Mol. Biol. 1993;22:517–23. doi: 10.1007/BF00015980. doi:10.1007/BF00015980. [DOI] [PubMed] [Google Scholar]
- 21.Chen W., Singh K.B. The auxin, hydrogen peroxide and salicylic acid induced expression of the Arabidopsis GST6 promoter is mediated in part by an ocs element. Plant J. 1999;19:667–77. doi: 10.1046/j.1365-313x.1999.00560.x. doi:10.1046/j.1365-313x.1999.00560.x. [DOI] [PubMed] [Google Scholar]
- 22.Brentner L.B., Mukherji S.T., Merchie K.M., Yoon J.M., Schnoor J.L., Van Aken B. Expression of glutathione S-transferases in poplar trees (Populus trichocarpa) exposed to 2,4,6-trinitrotoluene (TNT) Chemosphere. 2008;73:657–62. doi: 10.1016/j.chemosphere.2008.07.059. doi:10.1016/j.chemosphere.2008.07.059. [DOI] [PubMed] [Google Scholar]
- 23.Chen W., Chao G., Singh K.B. The promoter of a H2O2-inducible, Arabidopsis glutathione S-transferase gene contains closely linked OBF- and OBP1-binding sites. Plant J. 1996;10:955–66. doi: 10.1046/j.1365-313x.1996.10060955.x. doi:10.1046/j.1365-313X.1996.10060955.x. [DOI] [PubMed] [Google Scholar]
- 24.Jain M., Ghanashyam C., Bhattacharjee A. Comprehensive expression analysis suggests overlapping and specific roles of rice glutathione S-transferase genes during development and stress responses. BMC Genomics. 2010;11:73. doi: 10.1186/1471-2164-11-73. doi:10.1186/1471-2164-11-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Coleman J., Blake-Kalff M., Davies E. Detoxification of xenobiotics by plants: chemical modification and vacuolar compartmentation. Trends Plant Sci. 1997;2:144–51. doi:10.1016/S1360-1385(97)01019-4. [Google Scholar]
- 26.Roxas V.P., Smith R.K., Allen E.R., Allen R.D. Overexpression of glutathione S-transferase/glutathione peroxidase enhances the growth of transgenic tobacco seedlings during stress. Nat. Biotechnol. 1997;15:988–91. doi: 10.1038/nbt1097-988. doi:10.1038/nbt1097-988. [DOI] [PubMed] [Google Scholar]
- 27.Roxas V.P., Lodhi S.A., Garrett D.K., Mahan J.R., Allen R.D. Stress tolerance in transgenic tobacco seedlings that overexpress glutathione S-transferase/glutathione peroxidase. Plant Cell Physiol. 2000;41:1229–34. doi: 10.1093/pcp/pcd051. doi:10.1093/pcp/pcd051. [DOI] [PubMed] [Google Scholar]
- 28.Milligan A.S., Daly A., Parry M.A.J., Lazzeri P.A., Jepson I. The expression of a maize glutathione S-transferase in transgenic wheat confers herbicide tolerance, both in planta and in vivo. Mol. Breed. 2001;7:301–5. doi:10.1023/A:1011652821765. [Google Scholar]
- 29.Dixon D.P., McEwen A.G., Lapthorn A.J., Edwards R. Forced evolution of a herbicide detoxifying glutathione transferase. J. Biol. Chem. 2003;278:23930–5. doi: 10.1074/jbc.M303620200. doi:10.1074/jbc.M303620200. [DOI] [PubMed] [Google Scholar]
- 30.Yu T., Li Y.S., Chen X.F., Hu J., Chang X., Zhu Y.G. Transgenic tobacco plants overexpressing cotton glutathione S-transferase (GST) show enhanced resistance to methyl viologen. J. Plant Physiol. 2003;160:1305–11. doi: 10.1078/0176-1617-01205. doi:10.1078/0176-1617-01205. [DOI] [PubMed] [Google Scholar]
- 31.Light G.G., Mahan J.R., Roxas V.P., Allen R.D. Transgenic cotton (Gossypium hirsutum L.) seedlings expressing a tobacco glutathione S-transferase fail to provide improved stress tolerance. Planta. 2005;222:346–54. doi: 10.1007/s00425-005-1531-7. doi:10.1007/s00425-005-1531-7. [DOI] [PubMed] [Google Scholar]
- 32.Karavangeli M., Labrou N.E., Clonis Y.D., Tsaftaris A. Development of transgenic tobacco plants overexpressing maize glutathione S-transferase I for chloroacetanilide herbicides phytoremediation. Biomol. Eng. 2005;22:121–8. doi: 10.1016/j.bioeng.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 33.Marrs K.A., Alfenito M.R., Lloyd A.M., Walbot V. A glutathione S-transferase involved in vacuolar transfer encoded by the maize gene Bronze-2. Nature. 1995;375:397–400. doi: 10.1038/375397a0. doi:10.1038/375397a0. [DOI] [PubMed] [Google Scholar]
- 34.Alfenito M.R., Souer E., Goodman C.D., Buell R., Mol J., Koes R., Walbot V. Functional complementation of anthocyanin sequestration in the vacuole by widely divergent glutathione S-transferases. Plant Cell. 1998;10:1135–49. doi: 10.1105/tpc.10.7.1135. doi:10.1105/tpc.10.7.1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mueller L.A., Goodman C.D., Silady R.A., Walbot V. AN9, a petunia glutathione S-transferase required for anthocyanin sequestration, is a flavonoid-binding protein. Plant Physiol. 2000;123:1561–70. doi: 10.1104/pp.123.4.1561. doi:10.1104/pp.123.4.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kitamura S., Shikazono N., Tanaka A. TRANSPARENT TESTA 19 is involved in the accumulation of both anthocyanins and proanthocyanidins in Arabidopsis. Plant J. 2004;37:104–14. doi: 10.1046/j.1365-313x.2003.01943.x. doi:10.1046/j.1365-313X.2003.01943.x. [DOI] [PubMed] [Google Scholar]
- 37.Zettl R., Schell J., Palme K. Photoaffinity labeling of Arabidopsis thaliana plasma membrane vesicles by 5-azido-[7-3H]indole-3-acetic acid: identification of a glutathione S-transferase. Proc. Natl Acad. Sci. USA. 1994;91:689–93. doi: 10.1073/pnas.91.2.689. doi:10.1073/pnas.91.2.689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bilang J., Sturm A. Cloning and characterization of a glutathione S-transferase that can be photolabeled with 5-azidoindole-3-acetic acid. Plant Physiol. 1995;109:253–60. doi: 10.1104/pp.109.1.253. doi:10.1104/pp.109.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Smith A.P., Nourizadeh S.D., Peer W.A., Xu J., Bandyopadhyay A., Murphy A.S., Goldsbrough P.B. Arabidopsis At-GSTF2 is regulated by ethylene and auxin, and encodes a glutathione S-transferase that interacts with flavonoids. Plant J. 2003;36:433–42. doi: 10.1046/j.1365-313x.2003.01890.x. doi:10.1046/j.1365-313X.2003.01890.x. [DOI] [PubMed] [Google Scholar]
- 40.Gonneau M., Pagant S., Brun F., Laloue M. Photoaffinity labelling with the cytokinin agonist azido-CPPU of a 34 kDa peptide of the intracellular pathogenesis-related protein family in the moss Physcomitrella patens. Plant Mol. Biol. 2001;46:539–48. doi: 10.1023/a:1010693213437. doi:10.1023/A:1010693213437. [DOI] [PubMed] [Google Scholar]
- 41.Lederer B., Boger P. Binding and protection of porphyrins by glutathione S-transferases of Zea mays L. Biochim. Biophys. Acta. 2003;1621:226–33. doi: 10.1016/s0304-4165(03)00073-4. [DOI] [PubMed] [Google Scholar]
- 42.Nobuta K., Venu R.C., Lu C., et al. An expression atlas of rice mRNAs and small RNAs. Nat. Biotechnol. 2007;25:473–7. doi: 10.1038/nbt1291. doi:10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- 43.Kojima H., Suzuki T., Kato T., et al. Sugar-inducible expression of the nucleolin-1 gene of Arabidopsis thaliana and its role in ribosome synthesis, growth and development. Plant J. 2007;49:1053–63. doi: 10.1111/j.1365-313X.2006.03016.x. doi:10.1111/j.1365-313X.2006.03016.x. [DOI] [PubMed] [Google Scholar]
- 44.Jiang S.Y., Bachmann D., La H., Ma Z., Venkatesh P.N., Ramamoorthy R., Ramachandran S. Ds insertion mutagenesis as an efficient tool to produce diverse variations for rice breeding. Plant Mol. Biol. 2007;65:385–402. doi: 10.1007/s11103-007-9233-0. doi:10.1007/s11103-007-9233-0. [DOI] [PubMed] [Google Scholar]
- 45.Jiang S.Y., Christoffels A., Ramamoorthy R., Ramachandran S. Expansion mechanisms and functional annotations of hypothetical genes in the rice genome. Plant Physiol. 2009;150:1997–2008. doi: 10.1104/pp.109.139402. doi:10.1104/pp.109.139402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Matsui A., Ishida J., Morosawa T., et al. Arabidopsis transcriptome analysis under drought, cold, high-salinity and ABA treatment conditions using a tiling array. Plant Cell Physiol. 2008;49:1135–49. doi: 10.1093/pcp/pcn101. doi:10.1093/pcp/pcn101. [DOI] [PubMed] [Google Scholar]
- 47.Lin H., Ouyang S., Egan A., et al. Characterization of paralogous protein families in rice. BMC Plant Biol. 2008;8:18. doi: 10.1186/1471-2229-8-18. doi:10.1186/1471-2229-8-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Blanc G., Wolfe K.H. Functional divergence of duplicated genes formed by polyploidy during Arabidopsis evolution. Plant Cell. 2004;16:1679–91. doi: 10.1105/tpc.021410. doi:10.1105/tpc.021410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Gu Z., Nicolae D., Lu H.H., Li W.H. Rapid divergence in expression between duplicate genes inferred from microarray data. Trends Genet. 2002;18:609–13. doi: 10.1016/s0168-9525(02)02837-8. doi:10.1016/S0168-9525(02)02837-8. [DOI] [PubMed] [Google Scholar]
- 50.Finn R.D., Mistry J., Schuster-Böckler B., et al. Pfam: clans, web tools and services. Nucleic Acids Res. 2006;34:D247–51. doi: 10.1093/nar/gkj149. doi:10.1093/nar/gkj149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.G. The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–82. doi: 10.1093/nar/25.24.4876. doi:10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Letunic I., Doerks T., Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37(Database issue):D229–32. doi: 10.1093/nar/gkn808. doi:10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview version 2—a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–91. doi: 10.1093/bioinformatics/btp033. doi:10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Jiang S.Y., Ramachandran S. Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiol. Genomics. 2006;24:235–51. doi: 10.1152/physiolgenomics.00210.2005. [DOI] [PubMed] [Google Scholar]
- 55.Lehti-Shiu M.D., Zou C., Hanada K., Shiu S.H. Evolutionary history and stress regulation of plant receptor-like kinase/pelle genes. Plant Physiol. 2009;150:12–26. doi: 10.1104/pp.108.134353. doi:10.1104/pp.108.134353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Simillion C., Vandepoele K., Van Montagu M.C., Zabeau M., van de Peer Y. The hidden duplication past of Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2002;99:13627–32. doi: 10.1073/pnas.212522399. doi:10.1073/pnas.212522399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vandepoele K., Simillion C., Van de Peer Y. Evidence that rice and other cereals are ancient aneuploids. Plant Cell. 2003;15:2192–202. doi: 10.1105/tpc.014019. doi:10.1105/tpc.014019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lin H., Zhu W., Silva J.C., Gu X., Buell C.R. Intron gain and loss in segmentally duplicated genes in rice. Genome Biol. 2006;7:R41. doi: 10.1186/gb-2006-7-5-r41. doi:10.1186/gb-2006-7-5-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kong H., Landherr L.L., Frohlich M.W., Leebens-Mack J., Ma H., de Pamphilis C.W. Patterns of gene duplication in the plant SKP1 gene family in angiosperms: evidence for multiple mechanisms of rapid gene birth. Plant J. 2007;50:873–85. doi: 10.1111/j.1365-313X.2007.03097.x. doi:10.1111/j.1365-313X.2007.03097.x. [DOI] [PubMed] [Google Scholar]
- 60.Xu Z., Wang H. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35:D265–8. doi: 10.1093/nar/gkm286. doi:10.1093/nar/gkm286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Chaparro C., Guyot R., Zuccolo A., Piégu B., Panaud O. RetrOryza: a database of the rice LTR-retrotransposons. Nucleic Acids Res. 2007;35:D66–70. doi: 10.1093/nar/gkl780. doi:10.1093/nar/gkl780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Sheehan D., Meade G., Foley V.M., Dowd C.A. Structure, function and evolution of glutathione transferases: implications for classification of non-mammalian members of an ancient enzyme superfamily. Biochem. J. 2001;360:1–16. doi: 10.1042/0264-6021:3600001. doi:10.1042/0264-6021:3600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wagner U., Edwards R., Dixon D.P., Mauch F. Probing the diversity of the Arabidopsis glutathione S-transferase gene family. Plant Mol. Biol. 2002;49:515–32. doi: 10.1023/a:1015557300450. doi:10.1023/A:1015557300450. [DOI] [PubMed] [Google Scholar]
- 64.Shiu S.H., Karlowski W.M., Pan R., Tzeng Y.H., Mayer K.F., Li W.H. Comparative analysis of the receptor-like kinase family in Arabidopsis and rice. Plant Cell. 2004;16:1220–34. doi: 10.1105/tpc.020834. doi:10.1105/tpc.020834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiang S.Y., Ma Z., Ramachandran S. Evolutionary history and stress regulation of the lectin superfamily in higher plants. BMC Evol. Biol. 2010;10:79. doi: 10.1186/1471-2148-10-79. doi:10.1186/1471-2148-10-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.da Fonseca R.R., Johnson W.E., O'Brien S.J., Vasconcelos V., Antunes A. Molecular evolution and the role of oxidative stress in the expansion and functional diversification of cytosolic glutathione transferases. BMC Evol. Biol. 2010;10:281. doi: 10.1186/1471-2148-10-281. doi:10.1186/1471-2148-10-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Frova C. Glutathione transferases in the genomics era: New insights and perspectives. Biomol. Eng. 2006;23:149–69. doi: 10.1016/j.bioeng.2006.05.020. doi:10.1016/j.bioeng.2006.05.020. [DOI] [PubMed] [Google Scholar]
- 68.Koonin E.V., Mushegian A.R., Tatusov R.L., Altschul S.F., Bryant S.H., Bork P., Valencia A. Eukaryotic translation elongation factor 1c contains a glutathione transferase domain - Study of a diverse, ancient protein superfamily using motif search and structural modeling. Protein Sci. 1994;3:2045–54. doi: 10.1002/pro.5560031117. doi:10.1002/pro.5560031117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Martin J.L. Thioredoxin—a fold for all reasons. Structure. 1995;3:245–50. doi: 10.1016/s0969-2126(01)00154-x. doi:10.1016/S0969-2126(01)00154-X. [DOI] [PubMed] [Google Scholar]
- 70.Oakley A.J. Glutathione transferases: new functions. Curr. Opin. Struct. Biol. 2005;15:716–23. doi: 10.1016/j.sbi.2005.10.005. doi:10.1016/j.sbi.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 71.Apic G., Gough J., Teichmann S.A. Domain combinations in archaeal, eubacterial and eukaryotic proteomes. J. Mol. Biol. 2001;310:311–25. doi: 10.1006/jmbi.2001.4776. doi:10.1006/jmbi.2001.4776. [DOI] [PubMed] [Google Scholar]
- 72.Chothia C., Gough J., Vogel C., Teichmann S.A. Evolution of the protein repertoire. Science. 2003;300:1701–3. doi: 10.1126/science.1085371. doi:10.1126/science.1085371. [DOI] [PubMed] [Google Scholar]
- 73.Dixon D.P., Hawkins T., Hussey P.J., Edwards R. Enzyme activities and subcellular localization of members of the Arabidopsis glutathione transferase superfamily. J. Exp. Bot. 2009;60:1207–18. doi: 10.1093/jxb/ern365. doi:10.1093/jxb/ern365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Flagel L.E., Wendel J.F. Gene duplication and evolutionary novelty in plants. New Phytol. 2009;183:557–64. doi: 10.1111/j.1469-8137.2009.02923.x. doi:10.1111/j.1469-8137.2009.02923.x. [DOI] [PubMed] [Google Scholar]
- 75.Demuth J.P., Hahn M.W. The life and death of gene families. Bioessays. 2009;31:29–39. doi: 10.1002/bies.080085. doi:10.1002/bies.080085. [DOI] [PubMed] [Google Scholar]
- 76.Paterson A.H., Bowers J.E., Bruggmann R., et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–6. doi: 10.1038/nature07723. doi:10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 77.Bedell J.A., Budiman M.A., Nunberg A., et al. Sorghum genome sequencing by methylation filtration. PLoS Biol. 2005;3:e13. doi: 10.1371/journal.pbio.0030013. doi:10.1371/journal.pbio.0030013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Goff S.A., Ricke D., Lan T.H., et al. A draft sequence of the rice genome (Oryza sativa L. ssp. japonica) Science. 2002;296:92–100. doi: 10.1126/science.1068275. doi:10.1126/science.1068275. [DOI] [PubMed] [Google Scholar]
- 79.Juretic N., Hoen D.R., Huynh M.L., Harrison P.M., Bureau T.E. The evolutionary fate of MULE-mediated duplications of host gene fragments in rice. Genome Res. 2005;15:1292–7. doi: 10.1101/gr.4064205. doi:10.1101/gr.4064205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Gonzali S., Loreti E., Solfanelli C., Novi G., Alpi A., Perata P. Identification of sugar-modulated genes and evidence for in vivo sugar sensing in Arabidopsis. J. Plant Res. 2006;119:115–23. doi: 10.1007/s10265-005-0251-1. doi:10.1007/s10265-005-0251-1. [DOI] [PubMed] [Google Scholar]
- 81.Jiang S.Y., Ramamoorthy R., Ramachandran S. Comparative transcriptional profiling and evolutionary analysis of the GRAM domain family in eukaryotes. Dev. Biol. 2008;314:418–32. doi: 10.1016/j.ydbio.2007.11.031. doi:10.1016/j.ydbio.2007.11.031. [DOI] [PubMed] [Google Scholar]
- 82.Salzman R.A., Brady J.A., Finlayson S.A., et al. Transcriptional profiling of sorghum induced by methyl jasmonate, salicylic acid, and aminocyclopropane carboxylic acid reveals cooperative regulation and novel gene responses. Plant Physiol. 2005;138:352–68. doi: 10.1104/pp.104.058206. doi:10.1104/pp.104.058206. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.