Abstract
Two-component systems (TCSs) play vital functions in the adaptation of plants to environmental stresses. To identify soybean TCS genes involved in the regulation of drought stress response, we performed tissue-specific expression profiling of all 83 putative TCS genes in plants subjected to dehydration. Under well-watered conditions, the majority of soybean TCS genes were expressed higher in the root tissues. Additionally, a high variability in transcript abundance was observed for the TCS genes in both roots and shoots. Under dehydration, TCS genes were more responsive in shoots than in roots. Further analysis indicated that 50% more TCS genes were repressed by dehydration than induced. Specifically, 18 genes were induced by 2-fold or more, whereas 33 genes were down-regulated at least 2-fold by dehydration. TCS genes putatively involved in cytokinin and ethylene signallings strongly responded to dehydration, suggesting that crosstalk exists between different hormonal and stress pathways. Our study provides the first glance into the complex regulatory roles of soybean TCSs underlying their functions in response to dehydration. Additionally, these systematic expression analyses identified excellent dehydration-responsive candidate genes to further clarify soybean TCS functions in drought response and to enable the development of improved drought tolerance in transgenic soybeans.
Keywords: soybean, two-component system, dehydration, expression profiling, RT-qPCR
1. Introduction
Drought stress is one of the several adverse environmental factors that are commonly encountered by plants and can result in significant reductions in crop productivity worldwide. In response to drought stress, plants activate a number of endogenous defence mechanisms that function to increase the drought tolerance.1 Plants have evolved molecular machinery that initiates complex signal-transduction networks which minimize the impact of a suboptimal water supply on plants. The identification of signalling pathways acting in stress-affected cells and the mutual interactions between these pathways are the major research efforts. As a result of the complexities of stress-related signal transduction and its huge potential for impact on modern agriculture, this research field has garnered a substantial amount of attention. The early events of plant adaptation to environmental stresses involve the perception of stress signals and subsequent signal transduction, leading to the activation of various physiological and metabolic responses.1–4
In the signal-transduction networks involved in the perception of stress signals to stress-responsive gene expression, phosphorylation, which is mediated by two-component systems (TCSs) or His-to-Asp phosphorelays, is a key mechanism for stress signal transduction in cells. TCSs, consisting of sensor hybrid histidine kinases (HKs), histidine phosphotransfers (HPts) and effector response regulators (RRs), have been systematically identified and analysed in two completely sequenced and well-annotated model plant species: Arabidopsis thaliana and rice (Oryza sativa).5–7 A number of studies in Arabidopsis have suggested that among the Arabidopsis HKs (AHKs), AHK1, AHK2, AHK3 and AHK4 function in response to drought stress. In planta studies have demonstrated that the cytokinin (CK)-independent AHK1 functions as a positive regulator, whereas the CK-responsive AHK2, AHK3 and AHK4 function as negative regulators in drought stress signalling in both ABA-dependent and ABA-independent pathways.8,9 Although the AHK2, AHK3 and AHK4 proteins all positively regulate shoot growth, they exert a negative regulation of root growth.10–13 AHK1 also positively regulates shoot growth but its function in root growth is not known at this time.8 The involvement of AHKs in drought stress response suggests that the downstream Arabidopsis HPts (AHPs) and Arabidopsis RRs (ARRs) may also function in drought stress response. However, at the present time, there is not any in planta evidence or expression data, which have characterized the regulatory roles of AHPs in drought stress signalling. As for the ARRs, which can be classified into the type-A, type-B, type-C and pseudo ARRs based on their sequence signatures,14 loss-of-function studies indicated that among the type-A ARRs, ARR3, ARR4, ARR5 and ARR6 may function as positive regulators, whereas ARR8 and ARR9 as negative regulators in osmotic stress response, suggesting that these type-A ARRs might play a role in drought stress response.9 Recently, mutations in the pseudo aprr5, aprr7 and aprr9 genes enhanced tolerance of the triple mutant to drought stress, demonstrating that these three APRRs play negative roles in drought response.15 Since the rice TCS genes have been identified,7 expression studies have indicated that transcription of several rice TCS members, including HK, HPt and RR encoding genes, is altered by salt stress treatments.16–18 However, drought-related functions and/or expression profiling of rice TCS genes under dehydration and/or drought stress remain to be determined. TCSs were also identified in the important model legume Lotus japonicus.19 None of the abiotic stress-related data are currently available for L. japonicus TCS components.
Global soybean (Glycine max) production is frequently impacted by drought stress, which may reduce soybean yield by ∼40%.20 Given the importance of TCS signalling pathways in the regulation of various biological processes and responses to environmental stimuli, including drought stress, we have recently compiled a list of putative TCS-associated components in soybean.21 Within the soybean genome, a total of 21 HK, 10 authentic and 3 pseudo HPt, 18 type-A RR, 15 type-B RR, 3 type-C RR and 13 pseudo RR encoding genes were identified. Comparative analysis of the Arabidopsis, rice and soybean TCS members revealed the conserved architecture of the TCSs between these plants.21 In this study, we aimed to identify tissue-related and dehydration-related soybean TCS genes by characterizing their expression profiles in both root and shoot tissues of soybean plants treated with a time-coursed dehydration stress. These findings will enable us to perform in planta functional analyses of the candidates and will allow us to identify appropriate stress-responsive TCS candidate genes and their respective promoters for the future improvement of drought resistance in soybean via genetic engineering. Therefore, the identification, characterization and molecular tailoring of novel TCS members will have the potential to overcome a number of important limitations involved in the generation of transgenic soybean plants with superior yield under drought conditions.1,2,8
2. Materials and methods
2.1. Plant growth, dehydration treatment and collection of tissues
Soybean cv. Williams 82 seeds were germinated in 6-l pots containing vermiculite and were well-watered and grown under greenhouse conditions (continuous 30°C temperature, photoperiod of 12 h/12 h, 80 µmol m−2 s−1 photon flux density and 60% relative humidity). For tissue-specific expression profiling of TCS genes, root and shoot tissues were collected from 12-day-old soybean plants in three biological repeats. For expression profiling of TCS genes under dehydration stress, the dehydration treatment was carried out in time-course experiments to identify dynamic changes in transcripts in response to dehydration stress. Specifically, 12-day-old plants were carefully removed from soil, and roots were gently washed to remove soil. The plants were subsequently transferred onto a filter paper and allowed to dry for 2 and 10 h under the following conditions: 60% relative humidity, 25°C temperature and 10 µmol m−2 s−1 photon flux light intensity. The intensity of the dehydration stress treatments was quantified by the levels of relative water contents and standardized water contents, which were determined as described previously (n = 5; Supplementary Fig. S1).22 For mock control, seedlings were removed from soil and washed exactly as above, then grown hydroponically in water for 2 and 10 h under the same conditions. Root and shoot tissues of dehydrated and control plants were separately collected in three biological repeats for expression profiling.
2.2. RNA isolation, DNase treatment and cDNA synthesis
Plant tissue samples were ground into a fine powder using a mortar and pestle. Total RNA was isolated using the TRIZOL reagent (Invitrogen), and RNA concentration was determined using the NanoDrop ND-1000 UV-Vis spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA). For each sample, 4 µg of total RNA was digested according to the manufacturer's instructions in a 25-µl volume with Turbo DNA-free DNaseI to remove genomic DNA contamination (Ambion, Austin, TX, USA). After DNaseI treatment, RNA concentration was determined again with the NanoDrop spectrophotometer. The A260/A280 values of all RNA samples used in this study were 2.08 or higher, and the A260/A230 ratios of all samples were higher than 2.2. First-strand cDNA synthesis was performed using 1 µg of DNaseI-treated RNA with the ReverTra Ace® qPCR RT Kit (Toyobo, Japan) in a 20-µl reaction volume according to the manufacturer's supplied protocol.
2.3. Quantitative real-time PCR
Gene-specific primers for soybean TCS genes were designed using the Primer3 software.23 Primer specificity was confirmed by blasting each primer sequence against the soybean genome (Glyma1 model),24 followed by analysing the melting curves and amplicon fragments. Primers were redesigned if the corresponding melting curve did not yield a single sharp peak and/or if they had an electrophoresis pattern that failed to produce a single amplicon of the correct predicted length. The CYP2 gene was selected as a reference gene in the expression profiling of soybean genes as recommended previously.25 Quantitative real-time PCRs (RT-qPCR) were performed in 96-well plates on a Stratagene MX3000P system (Agilent Technologies, Santa Clara, CA, USA) using Thunderbird™ SYBR® qPCR Mix (Toyobo, Japan) reagents. Primer sets of 0.4 µM final concentrations for each primer were used in a final volume of 10 µl well−1. The thermal profile of the RT-qPCRs was at 95°C for 1 min, 40 cycles at 95°C for 15 s and at 60°C for 1 min. Dissociation curves were obtained using a thermal melting profile performed after the last PCR cycle: 95°C for 15 s followed by a constant increase in the temperature between 60°C and 95°C. Background-corrected raw fluorescence data were exported from the MX3000P system and analysed in LinRegPCR software with a built-in baseline correction and amplification efficiency calculation.26,27 Amplicon-based fluorescence thresholds were used to obtain the Ct values, and these values together with the amplicon-based mean efficiency were used for calculating the initial quantity of mRNA transcripts. Finally, the mRNA levels of each transcript were normalized with those of the corresponding CYP2 transcript.
2.4. Statistical analysis of the data
Mean values of three biological repeats were used to plot figures, and error bars on each figure represent the standard errors. When appropriate, a Student's t-test (one tail, unpaired, equal variance) was used to determine the statistical significance of the differential expression patterns between tissues and/or between treatments. Differential expression data which failed to pass the t-test with a P-value <0.05 were regarded as insignificant. These insignificant data were regarded as ‘ubiquitous’ in the case of tissue-specific comparisons and as ‘unresponsive’ in comparisons between normal and stress treatments; regardless of the fold change between the expression levels. Dehydration-responsive genes were defined as the differentially expressed genes with at least 2-fold induction or repression at 2 h and/or 10 h after dehydration treatment, whereas their expression in the mock control seedlings did not significantly change in a similar manner (Student's t-test, P < 0.05).
3. Results and discussion
3.1. Confirmation of primer specificity
Since the specificity of primers is crucial in RT-qPCR assays,28 we carefully designed each primer pair as described in the ‘Materials and methods' section. Furthermore, we determined their specificity by analysing their respective melting curves and amplicon fragments. All primer sets used in this study produced only one peak in their respective melting curves, and corresponding amplicons detected under our RT-qPCR conditions generated a single band of expected size on 2% agarose gels (data not shown). The sequences and calculated amplification efficiencies of the specific primers used in this study can be obtained in Supplementary Table S1.
3.2. Expression of the soybean TCS genes in root and shoot tissues
Increasing evidence suggests that the mechanisms of drought resistance are either related to root and/or shoot traits.29 Root morphology and development is one of the key traits that is correlated with mechanisms of drought resistance.30 Because the distribution of water within the rhizosphere is critical to maintaining function in different environmental conditions, plasticity for root traits is a vital factor to acquire resources. For instance, plants can adapt to drought by developing a longer taproot which helps reach lower soil layers where water is more readily available. In addition, an extensive fibrous root system can be useful for foraging subsoil surface moisture and nutrients such as phosphorus.29 On the other hand, a restraint of shoot growth has been shown to be advantageous in adverse environments by minimizing the evaporative leaf surface area. The growth restraint conferred by DELLA proteins, whose functions are regulated by different hormones such as gibberellin (GA), auxin and ethylene, is beneficial and promotes survival.31 Overexpression of the GhDREB1 gene from cotton (Gossypium hirsutum) in Arabidopsis resulted in a cold, salt and osmotic stress-tolerant phenotype as a consequence of growth retardation caused by GA deficiency, decreased sensitivity to CKs and repression of CK signalling.32 Therefore, the appropriate control of shoot- and root-related morphological traits is a promising approach for developing drought resistance in a number of crops, including soybean.29,31,32
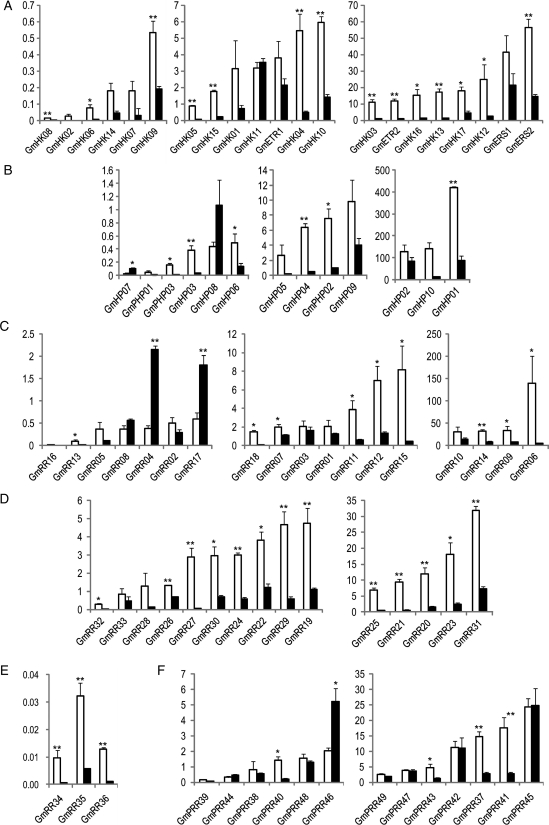
In Arabidopsis, numerous reports have suggested that TCS members regulate shoot and root growth and branching.10–14 In order to identify candidate genes that could be potentially used for enhancing drought resistance by altering shoot and/or root growth when overexpressed or repressed in transgenic plant systems, we determined expression profiles for all root- and shoot-specific soybean TCS members. As shown in Fig. 1 and summarized in Table 1 (detailed in Supplementary Table S2), the majority of soybean TCS genes were preferentially or specifically expressed in roots according to the criteria defined for the analysis of tissue-specific expression (Table 1; Supplementary Table S2).33 Forty-eight of the 83 soybean TCS genes were expressed highly in roots. Among these 48 genes, 20 genes were further classified into a root-specific group, as their transcript abundance in roots was more than 10-fold higher than that of shoots. The remaining 28 genes fell into the root-preferential group as their corresponding root/shoot expression ratios were between 3 and 10 (Fig. 1, Table 1). One gene (GmRR18) was found to be ‘very specifically expressed’ in root tissues as its root/shoot transcript abundance ratio was more than 118-fold (Fig. 1C). Another gene (GmHK02) was identified as a root ‘exclusively expressed’ gene since it was only detected in root tissues (Fig. 1A). Only four genes (GmHP07, GmRR04, GmRR17 and GmPRR46) displayed higher expression in shoots and the remaining 30 genes showed ubiquitous expression patterns. All four of the shoot-related genes were grouped into the shoot-preferential group with root/shoot expression ratios ranged between −10 and −3 (Fig. 1, Table 1, Supplementary Table S2).
Figure 1.
Expression of soybean TCS genes in root (white bars) and shoot (black bars) tissues under normal conditions. (A) GmHK genes. (B) GmHP and GmPHP genes. (C) Type-A GmRR genes. (D) Type-B GmRR genes. (E) Type-C GmRR genes. (F) Pseudo GmPRR genes. Data represent the means and standard errors of three independent biological samples. Asterisks on the top of bars indicate statistically significant differences between tissues with a P-value <0.05 (*) or 0.01 (**).
Table 1.
Expression of the soybean TCS genes in root and shoot tissues
| Fold differencea | Root-specific (X > 10-fold) | Root-preferential (3 < X < 10) | Ubiquitousb (−3 < X < 3) | Shoot-preferential (−10 < X < −3) | Shoot-specific (X < −10) |
|---|---|---|---|---|---|
| HKs | 8 | 7 | 6 | 0 | 0 |
| Phosphotransfer proteins | 3 | 3 | 6 | 1 | 0 |
| RRs | |||||
| Type-A | 3 | 5 | 7 | 2 | 0 |
| Type-B | 4 | 8 | 3 | 0 | 0 |
| Type-C | 2 | 1 | 0 | 0 | 0 |
| Pseudo | 0 | 4 | 8 | 1 | 0 |
| Total | 20 | 28 | 30 | 4 | 0 |
aFold difference was calculated as the ratio of mean expression levels of the same gene in roots and the shoots, when the root/shoot ratio is <1.0, the ratio was reversed and a minus sign (−) was added.
bGenes whose differential expressions between root and the shoot tissues did not pass Student's t-test with P-value <0.05 are also classified as ubiquitously expressed genes.
The expression levels of the soybean TCS genes were widely divergent. For example, the GmHK genes, which express more highly in roots than in shoots, can be divided into three groups (low, medium and high) based on their transcript abundance, in which the lowest (GmHK08) and highest (GmERS2) transcript abundance in roots was more than 3200-fold different (Fig. 1A). Among the GmHK genes, the genes encoding ethylene-receptor HKs, especially the GmERS1 and GmERS2, were expressed more highly than the others in both root and shoot tissues. Three GmHK genes (GmHK07–09), encoding proteins with highest homology to the osmosensor AHK1 of Arabidopsis,21 have a similar root-enriched expression pattern to the AHK1 gene.34 Similarly, TCS genes, coding for the Arabidopsis AHK4-like HKs in soybean (GmHK14–17), are more preferentially expressed in roots than in shoots (Fig. 1A),10,21 suggesting that a correlation between sequence conservation and expression patterns may exist. The soybean HPt encoding TCS genes formed a highly divergent group based on their transcript abundance. Specifically, their highest/lowest expression ratios in root and shoot tissues were more than 16 000- and 65 000-fold, respectively (Fig. 1B). Among the GmRR genes, the type-A GmRRs possessed the most diverse expression levels in both roots and shoots (Fig. 1C). GmRR06 had the highest expression in the root tissues, which was 170 000-fold higher than that of the lowest one (GmRR16). The expression levels among type-A GmRR genes in shoots were less variable than in roots, and the difference in expression was 28 000-fold between the highest (GmRR10) and the lowest (GmRR16) expressed genes (Fig. 1C). Twelve of 15 genes encoding type-B GmRRs were expressed either preferentially (eight genes) or specifically (four genes) in root tissues (Table 1). There were no shoot-preferential expression patterns detected among the type-B GmRR genes (Fig. 1D). The greatest amount of divergence in root/shoot expression levels among GmRRs was found for GmRR27, which had a root/shoot transcript abundance ratio of 39-fold. According to a comparative sequence analysis,21 the 15 type-B GmRRs showed the highest amino acid sequence identity to five ARRs (ARR1, ARR2, ARR11, ARR12 and ARR14). With the exception of the ARR14 gene that was not expressed in roots, semi-quantitative analyses determined that the remaining four ARR genes were expressed ubiquitously in root and leaf tissues.35 All of the GmPRR genes were expressed in both root and shoot tissues, of which four, eight and one genes were expressed root preferentially, ubiquitously and shoot preferentially, respectively (Table 1, Fig. 1F). Among the soybean TCS genes, the three type-C GmRR genes (GmRR34–36) exhibited the lowest levels of expression under our experimental conditions (Fig. 1E). In an analysis of soybean short transcript sequence reads by Libault et al.,33 the transcripts of two type-C GmRR genes (GmRR34 and GmRR35) were not detected in all the tissues except from the green pods. It should also be noted that the transcripts of GmHK05, GmHK06 and GmRR36 were not found in any of the nine tissues tested in their studies, including a transcriptomic analysis of root hair cells.33,36 However, in our experimental settings, the presence of the transcripts of these genes were reliably detected, albeit at low levels (Fig. 1A and E). It is possible that this discrepancy is due to the duplicated nature of the soybean genome,37 resulting in very high homology between duplicates. As a result, the method used in the authors’ study failed to detect expression because it could not assign reads matching with two or more closely related loci.33,36
3.3. Expression of the soybean TCS gene during dehydration
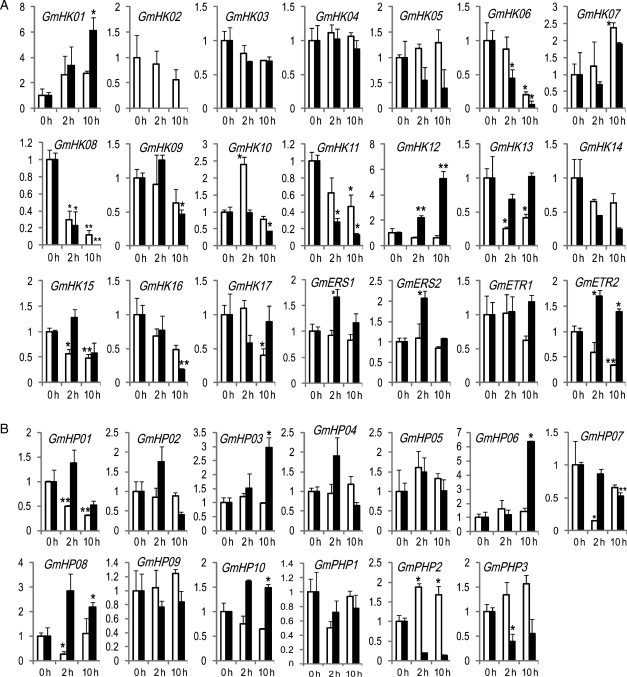
Plant TCSs have been implicated in the regulation of environmental stress responses, including drought stress,8,16–18,38 suggesting that the TCS genes, which function in stress responses, may be used to enhance drought tolerance in plants via genetic engineering.8,38 Stress-related soybean TCS genes can be predicted based on comparative sequence analyses21; however, this approach has limitations. Although TCSs have been systematically identified and characterized in Arabidopsis, rice, L. japonicus and soybean,5–7,19 only a few members of Arabidopsis TCS genes have been identified as drought-related genes based on expression and/or functional analyses.8,38 Cis-element-based targeted gene finding approach can also be used to predict stress-responsive genes, and tissue-specific genes.21,39–41 However, because of their short length (5- to 9-bp core) and flexibility, the frequency of a cis-motif sequence in the whole genome is relatively high. In addition, a number of cis-element sequences might be syntactically correct without providing practical regulatory function.42 Therefore, our major goal of this study was to characterize the expression profile of all soybean TCS members under dehydration stress in order to precisely identify dehydration-responsive TCS genes that can potentially used to engineer soybean plants with improved drought resistance. We employed RT-qPCR analyses to conduct systematic expression profiling of all 83 TCS genes in root and shoot tissues of 12-day-old soybean plants subjected to 2 and 10 h dehydration stress. The evaluation of expression patterns in individual stressed tissues, rather than whole plants, might provide information on the mode of action of stress-responsive genes in specific tissues.43 As presented in Figs 2 and 3, a large number of soybean TCS genes appeared to be dehydration responsive. To precisely determine the dehydration-responsive genes, the expression of those soybean TCS genes, whose mRNA levels were found to be changed under dehydration treatment, was also examined in the water-treated control plant samples (Supplementary Figs S2 and S3). As a result, among 83 soybean TCS genes, a total of 18 induced and 33 repressed genes were identified (Fig. 4, Table 2, Supplementary Table S3). Previously, a cis-motif-based prediction using 12 stress-responsive cis-motifs suggested that out of 83 soybean TCS genes, 30 genes might be dehydration responsive because these genes were found to contain dehydration-responsive ABRE and/or MYBR and/or MYCR motifs in their promoter regions.21 It should be noticed that the ABRE and MYBR were discovered as dehydration-inducible cis-motifs, whereas MYCR was shown to act as both dehydration-inducible and dehydration-repressible regulatory motifs.44 We found that among the 30 predicted dehydration-responsive TCS genes, 11 genes were confirmed by our RT-qPCR analysis. Eight genes (GmHK10, GmRR01, GmRR02, GmRR25, GmRR34, GmRR35, GmRR36 and GmPRR39) are induced, and three genes (GmHP01, GmRR09 and GmRR29), which contain MYCR(s) in their promoter regions, are repressed (Supplementary Table S3).21 Thus, although the cis-element-based targeted gene finding approach has demonstrated a wide application in the genome-wide prediction due to the availability of a large number of cis-elements in many plant species, expression profiles of the cis-motif-based predicted genes should be verified experimentally using an expression profiling approach, such as RT-qPCR, prior to the launching of laborious in planta functional studies.39,40,42,45
Figure 2.
Expression of TCS genes encoding HKs and HPts in root (white bars) and shoot (black bars) tissues of soybean plants under dehydration stress. (A) Expression of TCS genes encoding HKs. (B) Expression of TCS genes encoding HPts. Relative gene expression levels were normalized to a value at 1 in the untreated plant samples (0 h). Data represent the means and standard errors of three independent biological samples. Asterisks on the top of bars indicate statistically significant differences when compared with 0 h with a P-value <0.05 (*) or 0.01 (**).
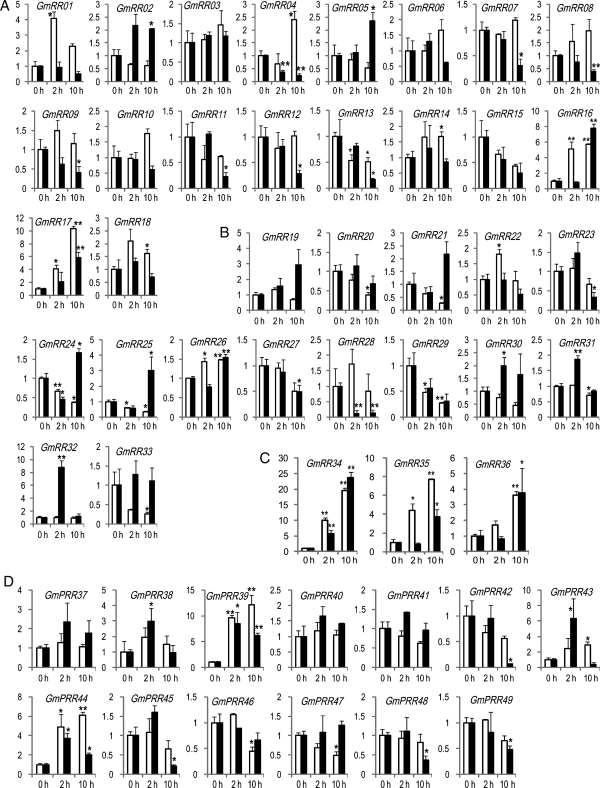
Figure 3.
Expression of TCS genes encoding RRs in root (white bars) and shoot (black bars) tissues of soybean plants under dehydration stress. (A) Type-A GmRR genes. (B) Type-B GmRR genes. (C) Type-C GmRR genes. (D) Pseudo GmPRR genes. Relative gene expression levels were normalized to a value at 1 in the untreated plant samples (0 h). Data represent the means and standard errors of three independent biological samples. Asterisks on the top of bars indicate statistically significant differences when compared with 0 h with a P-value <0.05 (*) or 0.01 (**).
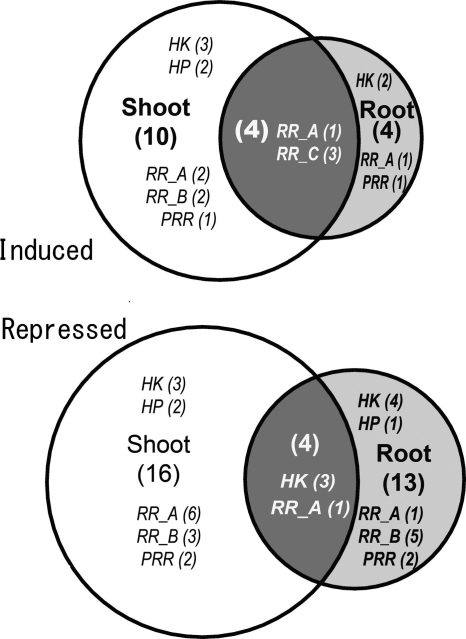
Figure 4.
Venn diagram showing dehydration-responsive soybean TCS genes in root and shoot tissues of soybean plants. The dehydration-responsive genes were defined as those genes whose expression is either induced (upper panel) or repressed (lower panel) significantly (P < 0.05) at least 2-fold at 2 h and/or 10 h after exposure to dehydration stress and their expression in the water-treated mock control samples did not significantly change in a similar manner. The reported differential expression patterns passed the Student's t-test (one tail, unpaired, assuming equal variance) with a P-value <0.05.
Table 2.
Genes with altered expression under dehydration in each family
| Family | Number of genes | Induced genes | Repressed genes |
|---|---|---|---|
| HKs | 21 | 5 | 10 |
| Phosphotransfers | |||
| Authentic | 10 | 2 | 2 |
| Pseudo | 3 | 0 | 2 |
| RRs | |||
| Type-A | 18 | 4 | 7 |
| Type-B | 15 | 2 | 8 |
| Type-C | 3 | 3 | 0 |
| Pseudo | 13 | 2 | 4 |
| Total genes | 83 | 18 | 33 |
Although the soybean TCS genes were predominantly expressed in roots than in shoots (Fig. 1), their expression patterns upon dehydration stress were opposite (Fig. 4). Specifically, there were only 8 and 17 genes whose expression was induced or repressed in the roots, respectively, upon dehydration. At the same time, there were 14 and 20 genes whose expression was either induced or repressed, respectively, in shoots (Fig. 4). Only a small number of the genes responded in a similar fashion (induced or repressed) in both tissues. Specifically, the GmHK06, GmHK08 and GmHK11 genes were down-regulated, and the GmRR16, GmRR34, GmRR35 and GmRR36 genes were up-regulated. These data support the hypothesis that the majority of soybean TCS genes respond to dehydration stress in either root- or shoot-specific manner. It is worth mentioning that the dehydration responsiveness of the GmHK10 gene were opposite in the root and the shoot tissues. The expression of GmHK10 was induced in the roots at 2 h but was repressed in the shoots at 10 h after dehydration treatment (Fig. 2A, Supplementary Fig. S2A). Not only having a larger number of dehydration-responsive genes, but the extent of the responsiveness of GmHKs, i.e. the fold change, was also higher in shoot tissues. For example, the expression levels of the GmHK01 and GmHK12 genes, which encode proteins most similar to the Arabidopsis CKI1 and AHK3, respectively,21 exhibited a 5–6-fold induction in shoots but insignificant induction in roots (Supplementary Table S3). Similarly, the extent of repression was also more severe in shoots than in roots. A maximum of 6-fold repression was found in roots, whereas a 17-fold repression was observed in shoots (Supplementary Table S3). GmHK08 was the most repressed among all soybean TCS genes during dehydration treatment and in both tissues, and its expression in shoots at 10 h after dehydration treatment was not detectable (Fig. 2A, Supplementary Fig. S2A). In Arabidopsis, the osmosensor AHK1 and the CK receptor AHK2, AHK3 and AHK4 have been shown to function in drought stress response. The expression of all four of these AHK genes is induced by dehydration.8 Among the three AHK1-like protein encoding GmHK07–09 genes,21 the expression of GmHK07 was induced by dehydration, suggesting that GmHK07 may play a positive regulatory role in drought response of soybean plants similar to that of AHK1 in Arabidopsis.8 Out of eight GmHK genes (GmHK10–17), which are predicted to encode CK-receptor HKs based on sequence similarity to their Arabidopsis counterparts,21 GmHK10 and GmHK12 showed significant induction in roots and shoots, respectively (Fig. 2A, Supplementary Fig. S2A). It is possible that GmHK10 and GmHK12 may act as negative regulators in drought stress response in a similar fashion as their Arabidopsis orthologs.8 Based on our analyses, all of the GmHK07, GmHK10 and GmHK12 genes appear to be good candidates for in planta studies.
Among the 13 soybean TCS genes encoding HPt proteins, two genes (GmHP03 and GmHP06) were induced and four other genes were repressed upon dehydration (Table 2). Interestingly, none of the induced genes were found in root tissues. Among the induced genes, GmHP06 exhibited the highest induction (>6-fold) in shoots, meanwhile its induction in roots was not significant. The most repressed gene was GmPHP2, whose expression was reduced by 7.5-fold in shoots upon dehydration (Fig. 2B, Supplementary Fig. S2B, Supplementary Table S3).
The expression of soybean GmRR genes followed different patterns (Fig. 3A, Supplementary Fig. S3, Supplementary Table S3). Repression of the type-A GmRRs genes in roots was not statistically significant. In addition, only two genes (GmRR01 and GmRR16) were induced in the same tissues. In shoots, three and six genes of this group were induced and repressed upon dehydration, respectively. Together, 11 of the 18 type-A GmRR genes were either induced and/or repressed by dehydration in root and/or shoot tissues, suggesting their diverse functions in the regulation of dehydration stress response (Supplementary Table S3). Diverse functions were also reported for Arabidopsis type-A ARRs. Mutations in arr3, arr4, arr5 and arr6 increase sensitivity, but an additional loss of ARR8 and ARR9 decreases the sensitivity to osmotic stress. These data suggest that ARR3, ARR4, ARR5 and ARR6 may function as positive regulators, whereas ARR8 and ARR9 function as negative regulators.9 In soybean, the six dehydration-repressed type-A GmRR genes (GmRR07–09 and GmRR11–13) encode ARR8- and ARR9-like GmRRs, providing correlative evidence that these GmRRs function in stress response, and may act as negative regulators in a similar fashion as their ARR8 and ARR9 orthologues (Supplementary Table S3). Similarly, the ARR4- and ARR6-like protein encoding GmRR01 and GmRR02 genes were up-regulated in response to drought, suggesting that they may function as positive regulators in stress response in a similar fashion as their Arabidopsis counterparts. The expression of type-B GmRR genes in soybean, which encode transcription factors, was mostly repressed in both tissues. Only two genes (GmRR25 and GmRR32) were induced in response to drought, and they exhibited shoot specificity (Fig. 3B, Table 2, Supplementary Fig. S3, Supplementary Table S3). As for the type-C GmRR genes, all three genes (GmRR34–36) were found to be induced strongly in both roots and shoots by dehydration (Fig. 3C, Table 2, Supplementary Fig. S3, Supplementary Table S3).
Among 13 pseudo GmPRR genes, the expression of six genes was altered by dehydration, suggesting that they may play a physiological role in response to dehydration (Fig. 3D, Table 2, Supplementary Fig. S3, Supplementary Table S3). Genes encoding GmPRR39 and GmPRR44 were significantly induced in shoots and roots, respectively, upon dehydration treatment. Four other genes encoding GmPRRs were repressed upon dehydration, of which two genes (GmPRR46 and GmPRR47) were repressed in the roots, whereas two other genes (GmPRR48 and GmPRR49) were repressed in the shoots (Supplementary Table S3). The dehydration-inducible GmPRR39 and GmPRR44 genes were shown to encode either APRR5- or APPR9-like proteins.21 In Arabidopsis, the APRR5 and APRR9 proteins were reported to negatively regulate drought response.15 Therefore, it is possible that the dehydration-responsive GmPRR39 and GmPRR44 may function as negative regulators in soybean as well. On the other hand, although the transcripts of GmPRR38, GmPRR42, GmPRR43 and GmPRR45 were accumulated under dehydration, the transcripts of these genes were also increased in a similar manner in the water-treated mock control plant tissues (Supplementary Fig. S3), suggesting that circadian clock-related regulation affected the expression of these GmPRR genes rather than dehydration stress. This is not a surprise because it has been reported previously that these four GmPRR genes encode circadian clock-associated APRR1-, APRR7- and APRR9-orthologous proteins.21,46
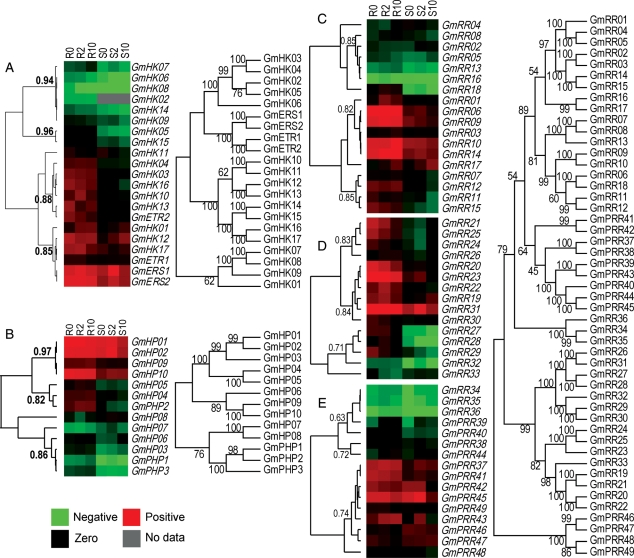
3.4. Clustering analysis of the expression data of soybean TCS genes
A gene's expression is regulated through several mechanisms, of which some are still unknown.47 Nonetheless, the regulation of gene expression via the interaction of transcription factors and promoters has been well documented.1 Similarities in expression patterns and responsiveness to stresses may likely be a result of the involvement of similar cis-elements and/or transcription factors. In order to gain an overall understanding of the expression patterns of soybean TCS genes, we performed hierarchical clustering using their log-transformed expression data. Figure 5 shows the result of hierarchical analysis of soybean TCS genes performed on each of the groups (GmHKs, GmHPs and GmRRs). As shown in Fig. 5A, the expression patterns of GmHK genes clustered into two distinct nodes. Within these two distinct nodes, each group was separated into two subnodes. The high correlation coefficient (0.85–0.96) of each subnode indicates that the expression patterns of the genes in each node were highly similar to one another. The expression of several genes with high homology, which is an indication of duplicated pairs, did not cluster in the analysis. For instance, GmHK02 and GmHK05, and GmHK10 and GmHK11 did not cluster, implying that their divergence in expression was the cause. The expression patterns of GmHP genes in roots and shoots during dehydration were clustered into three nodes with high correlation coefficient values (Fig. 5B). It should be noted that GmHP07 and GmHP08 did not fall into those nodes. With the exception of GmPHP1 and GmPHP2, the expressions of other highly homologous pairs in this group clustered together. These data indicated that functional redundancy is still high among duplicated HPt proteins of soybean. Expression profiles of each of the RR types (A, B, C and pseudo) clustered into two nodes mainly based on the expression levels (Fig. 5C–E). Some of the groups were clustered into two or more subgroups based on the responsiveness to dehydration in the tested tissues (Fig. 5C–E).
Figure 5.
Hierarchical clustering and heat map presentation for the expression of TCS genes in dehydrated root and shoot tissues of soybean. Genes are grouped according to their expression patterns using hierarchical clustering (Cluster v.2.11).48 The vertical dendrogram indicates the relationship among transcripts across tissues and treatments in the hierarchical clustering analysis. TreeView (http://rana.lbl.gov/eisen/?page_id=42) was used to generate the heat map figure. Phylogenetic trees are indicated on the right of each group. Numbers next to the nodes of the dendrogram indicate the correlation coefficient r. (A) HK proteins. (B) HPt proteins. (C) Type-A GmRR proteins. (D) Type-B GmRR proteins. (E) Type-C and pseudo GmRR proteins.
To find out how the soybean TCS genes cluster in response to dehydration, we conducted a hierarchical analysis based on the log-transformed ratio of the expression values with and without stress treatment. The groups of genes clustered in this analysis will not take into account the gene-to-gene expression divergence. We found that several nodes were formed (Supplementary Fig. S4), implying the possibility that similar dehydration-regulated pathways exist for each node.
3.5. Conclusions
This report has provided the first insight into the previously uncharacterized TCS members of soybean and placed a special emphasis on the relation to dehydration stress responsiveness. Our results have provided useful information by identifying candidate dehydration-responsive genes. By combining these genes with their associated dehydration-responsive promoters, scientists can utilize these resources to engineer soybean plants for enhanced stress resistance. We have selected a number of dehydration-responsive genes for further analysis in Arabidopsis to corroborate their functional significance in planta. By implementing these functional studies, we aim to identify a suite of candidate genes that are best suited for the genetic engineering of soybean plants with improved drought resistance.
Supplementary Data
Supplementary data are available at www.dnaresearch.oxfordjournals.org.
Funding
D.T.L. is supported by the RIKEN Foreign Postdoctoral Fellowship. This work was funded by a Start-up Support grant (M36-57000) from the RIKEN Yokohama Institute Director Discretionary Funds to L.-S.P.T.
References
- 1.Yamaguchi-Shinozaki K., Shinozaki K. Transcriptional regulatory networks in cellular responses and tolerance to dehydration and cold stresses. Annu. Rev. Plant Biol. 2006;57:781–803. doi: 10.1146/annurev.arplant.57.032905.105444. doi:10.1146/annurev.arplant.57.032905.105444. [DOI] [PubMed] [Google Scholar]
- 2.Valliyodan B., Nguyen H.T. Understanding regulatory networks and engineering for enhanced drought tolerance in plants. Curr. Opin. Plant Biol. 2006;9:189–95. doi: 10.1016/j.pbi.2006.01.019. doi:10.1016/j.pbi.2006.01.019. [DOI] [PubMed] [Google Scholar]
- 3.Tran L.S., Nakashima K., Shinozaki K., Yamaguchi-Shinozaki K. Plant gene networks in osmotic stress response: from genes to regulatory networks. Methods Enzymol. 2007;428:109–28. doi: 10.1016/S0076-6879(07)28006-1. doi:10.1016/S0076-6879(07)28006-1. [DOI] [PubMed] [Google Scholar]
- 4.Tran L.S., Nakashima K., Sakuma Y., et al. Co-expression of the stress-inducible zinc finger homeodomain ZFHD1 and NAC transcription factors enhances expression of the ERD1 gene in Arabidopsis. Plant J. 2007;49:46–63. doi: 10.1111/j.1365-313X.2006.02932.x. doi:10.1111/j.1365-313X.2006.02932.x. [DOI] [PubMed] [Google Scholar]
- 5.Hwang I., Chen H.C., Sheen J. Two-component signal transduction pathways in Arabidopsis. Plant Physiol. 2002;129:500–15. doi: 10.1104/pp.005504. doi:10.1104/pp.005504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ito Y., Kurata N. Identification and characterization of cytokinin-signalling gene families in rice. Gene. 2006;382:57–65. doi: 10.1016/j.gene.2006.06.020. doi:10.1016/j.gene.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 7.Pareek A., Singh A., Kumar M., Kushwaha H.R., Lynn A.M., Singla-Pareek S.L. Whole-genome analysis of Oryza sativa reveals similar architecture of two-component signaling machinery with Arabidopsis. Plant Physiol. 2006;142:380–97. doi: 10.1104/pp.106.086371. doi:10.1104/pp.106.086371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tran L.S., Urao T., Qin F., et al. Functional analysis of AHK1/ATHK1 and cytokinin receptor histidine kinases in response to abscisic acid, drought, and salt stress in Arabidopsis. Proc. Natl Acad. Sci. USA. 2007;104:20623–8. doi: 10.1073/pnas.0706547105. doi:10.1073/pnas.0706547105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wohlbach D.J., Quirino B.F., Sussman M.R. Analysis of the Arabidopsis histidine kinase ATHK1 reveals a connection between vegetative osmotic stress sensing and seed maturation. Plant Cell. 2008;20:1101–17. doi: 10.1105/tpc.107.055871. doi:10.1105/tpc.107.055871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Higuchi M., Pischke M.S., Mahonen A.P., et al. In planta functions of the Arabidopsis cytokinin receptor family. Proc. Natl Acad. Sci. USA. 2004;101:8821–6. doi: 10.1073/pnas.0402887101. doi:10.1073/pnas.0402887101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Werner T., Schmulling T. Cytokinin action in plant development. Curr. Opin. Plant Biol. 2009;12:527–38. doi: 10.1016/j.pbi.2009.07.002. doi:10.1016/j.pbi.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 12.Nishimura C., Ohashi Y., Sato S., Kato T., Tabata S., Ueguchi C. Histidine kinase homologs that act as cytokinin receptors possess overlapping functions in the regulation of shoot and root growth in Arabidopsis. Plant Cell. 2004;16:1365–77. doi: 10.1105/tpc.021477. doi:10.1105/tpc.021477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Riefler M., Novak O., Strnad M., Schmulling T. Arabidopsis cytokinin receptor mutants reveal functions in shoot growth, leaf senescence, seed size, germination, root development, and cytokinin metabolism. Plant Cell. 2006;18:40–54. doi: 10.1105/tpc.105.037796. doi:10.1105/tpc.105.037796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schaller G.E., Kieber J.J., Shiu S.-H. Two-component signaling elements and histidyl-aspartyl phosphorelays. In: Somerville C., Meyerowitz E., editors. The Arabidopsis Book. The American Society of Plant Biologists: Rockville, MD, USA; 2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nakamichi N., Kusano M., Fukushima A., et al. Transcript profiling of an Arabidopsis pseudo response regulator arrhythmic triple mutant reveals a role for the circadian clock in cold stress response. Plant Cell Physiol. 2009;50:447–62. doi: 10.1093/pcp/pcp004. doi:10.1093/pcp/pcp004. [DOI] [PubMed] [Google Scholar]
- 16.Jain M., Tyagi A.K., Khurana J.P. Molecular characterization and differential expression of cytokinin-responsive type-A response regulators in rice (Oryza sativa) BMC Plant Biol. 2006;6:1–2. doi: 10.1186/1471-2229-6-1. doi:10.1186/1471-2229-6-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jain M., Tyagi A.K., Khurana J.P. Differential gene expression of rice two-component signaling elements during reproductive development and regulation by abiotic stress. Funct. Integr. Genomics. 2008;8:175–80. doi: 10.1007/s10142-007-0063-6. doi:10.1007/s10142-007-0063-6. [DOI] [PubMed] [Google Scholar]
- 18.Karan R., Singla-Pareek S.L., Pareek A. Histidine kinase and response regulator genes as they relate to salinity tolerance in rice. Funct. Integr. Genomics. 2009;9:411–7. doi: 10.1007/s10142-009-0119-x. doi:10.1007/s10142-009-0119-x. [DOI] [PubMed] [Google Scholar]
- 19.Ishida K., Niwa Y., Yamashino T., Mizuno T. A genome-wide compilation of the two-component systems in Lotus japonicus. DNA Res. 2009;16:237–47. doi: 10.1093/dnares/dsp012. doi:10.1093/dnares/dsp012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tran L. S., Mochida K. Functional genomics of soybean for improvement of productivity in adverse conditions. Funct. Integr. Genomics. 2010;10:447–62. doi: 10.1007/s10142-010-0178-z. [DOI] [PubMed] [Google Scholar]
- 21.Mochida K., Yoshida T., Sakurai T., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. Genome-wide analysis of two-component systems and prediction of stress-responsive two-component system members in soybean. DNA Res. 2010;17:303–24. doi: 10.1093/dnares/dsq021. doi:10.1093/dnares/dsq021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fujita Y., Fujita M., Satoh R., et al. AREB1 Is a transcription activator of novel ABRE-dependent ABA signaling that enhances drought stress tolerance in Arabidopsis. Plant Cell. 2005;17:3470–88. doi: 10.1105/tpc.105.035659. doi:10.1105/tpc.105.035659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–86. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 24.Schmutz J., Cannon S.B., Schlueter J., et al. Genome sequence of the palaeopolyploid soybean. Nature. 2010;463:178–83. doi: 10.1038/nature08670. doi:10.1038/nature08670. [DOI] [PubMed] [Google Scholar]
- 25.Tran L.S., Quach T.N., Guttikonda S.K., et al. Molecular characterization of stress-inducible GmNAC genes in soybean. Mol. Genet. Genomics. 2009;281:647–64. doi: 10.1007/s00438-009-0436-8. doi:10.1007/s00438-009-0436-8. [DOI] [PubMed] [Google Scholar]
- 26.Ruijter J.M., Ramakers C., Hoogaars W.M., et al. Amplification efficiency: linking baseline and bias in the analysis of quantitative PCR data. Nucleic Acids Res. 2009;37:e45. doi: 10.1093/nar/gkp045. doi:10.1093/nar/gkp045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ramakers C., Ruijter J.M., Deprez R.H., Moorman A.F. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003;339:62–6. doi: 10.1016/s0304-3940(02)01423-4. doi:10.1016/S0304-3940(02)01423-4. [DOI] [PubMed] [Google Scholar]
- 28.Udvardi M.K., Czechowski T., Scheible W.R. Eleven golden rules of quantitative RT-PCR. Plant Cell. 2008;20:1736–37. doi: 10.1105/tpc.108.061143. doi:10.1105/tpc.108.061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manavalan L.P., Guttikonda S.K., Tran L.S., Nguyen H.T. Physiological and molecular approaches to improve drought resistance in soybean. Plant Cell Physiol. 2009;50:1260–76. doi: 10.1093/pcp/pcp082. doi:10.1093/pcp/pcp082. [DOI] [PubMed] [Google Scholar]
- 30.Sharp R.E., Poroyko V., Hejlek L.G., et al. Root growth maintenance during water deficits: physiology to functional genomics. J. Exp. Bot. 2004;55:2343–51. doi: 10.1093/jxb/erh276. doi:10.1093/jxb/erh276. [DOI] [PubMed] [Google Scholar]
- 31.Achard P., Cheng H., De Grauwe L., et al. Integration of plant responses to environmentally activated phytohormonal signals. Science. 2006;311:91–4. doi: 10.1126/science.1118642. doi:10.1126/science.1118642. [DOI] [PubMed] [Google Scholar]
- 32.Huang J.G., Yang M., Liu P., Yang G.D., Wu C.A., Zheng C.C. GhDREB1 enhances abiotic stress tolerance, delays GA-mediated development and represses cytokinin signalling in transgenic Arabidopsis. Plant Cell Environ. 2009;32:1132–45. doi: 10.1111/j.1365-3040.2009.01995.x. doi:10.1111/j.1365-3040.2009.01995.x. [DOI] [PubMed] [Google Scholar]
- 33.Libault M., Farmer A., Joshi T., et al. An integrated transcriptome atlas of the crop model Glycine max, and its use in comparative analyses in plants. Plant J. 2010;63:86–99. doi: 10.1111/j.1365-313X.2010.04222.x. [DOI] [PubMed] [Google Scholar]
- 34.Urao T., Yakubov B., Satoh R., et al. A transmembrane hybrid-type histidine kinase in Arabidopsis functions as an osmosensor. Plant Cell. 1999;11:1743–54. doi: 10.1105/tpc.11.9.1743. doi:10.1105/tpc.11.9.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mason M.G., Li J., Mathews D.E., Kieber J.J., Schaller G.E. Type-B response regulators display overlapping expression patterns in Arabidopsis. Plant Physiol. 2004;135:927–37. doi: 10.1104/pp.103.038109. doi:10.1104/pp.103.038109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Libault M., Farmer A., Brechenmacher L., et al. Complete transcriptome of the soybean root hair cell, a single-cell model, and its alteration in response to Bradyrhizobium japonicum infection. Plant Physiol. 2010;152:541–52. doi: 10.1104/pp.109.148379. doi:10.1104/pp.109.148379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Schlueter J.A., Lin J.Y., Schlueter S.D., et al. Gene duplication and paleopolyploidy in soybean and the implications for whole genome sequencing. BMC Genomics. 2007;8:330. doi: 10.1186/1471-2164-8-330. doi:10.1186/1471-2164-8-330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jeon J., Kim N.Y., Kim S., et al. A subset of cytokinin two-component signaling system plays a role in cold temperature stress response in Arabidopsis. J. Biol. Chem. 2010;285:23371–86. doi: 10.1074/jbc.M109.096644. doi:10.1074/jbc.M109.096644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang W., Ruan J., Ho T.H., You Y., Yu T., Quatrano R.S. Cis-regulatory element based targeted gene finding: genome-wide identification of abscisic acid- and abiotic stress-responsive genes in Arabidopsis thaliana. Bioinformatics. 2005;21:3074–81. doi: 10.1093/bioinformatics/bti490. doi:10.1093/bioinformatics/bti490. [DOI] [PubMed] [Google Scholar]
- 40.Kim D.W., Lee S.H., Choi S.B., et al. Functional conservation of a root hair cell-specific cis-element in angiosperms with different root hair distribution patterns. Plant Cell. 2006;18:2958–70. doi: 10.1105/tpc.106.045229. doi:10.1105/tpc.106.045229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Won S.K., Lee Y.J., Lee H.Y., Heo Y.K., Cho M., Cho H.T. Cis-element- and transcriptome-based screening of root hair-specific genes and their functional characterization in Arabidopsis. Plant Physiol. 2009;150:1459–73. doi: 10.1104/pp.109.140905. doi:10.1104/pp.109.140905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carroll S.B., Grenier J.K., Weatherbee S.D. From DNA to Diversity: Molecular Genetics and the Evolution of Animal Design. 1st edition. Malden, MA: Blackwell Science; 2001. p. p.214. [Google Scholar]
- 43.Tran L.S., Mochida K. Identification and prediction of abiotic stress responsive transcription factors involved in abiotic stress signaling in soybean. Plant Signal. Behav. 2010;5:255–7. doi: 10.4161/psb.5.3.10550. doi:10.4161/psb.5.3.10550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Abe H., Yamaguchi-Shinozaki K., Urao T., Iwasaki T., Hosokawa D., Shinozaki K. Role of Arabidopsis MYC and MYB homologs in drought- and abscisic acid-regulated gene expression. Plant Cell. 1997;9:1859–68. doi: 10.1105/tpc.9.10.1859. doi:10.1105/tpc.9.10.1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mochida K., Yoshida T., Sakurai T., Yamaguchi-Shinozaki K., Shinozaki K., Tran L.S. In silico analysis of transcription factor repertoire and prediction of stress responsive transcription factors in soybean. DNA Res. 2009;16:353–69. doi: 10.1093/dnares/dsp023. doi:10.1093/dnares/dsp023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ono N., Ishida K., Yamashino T., et al. Genomewide characterization of the light-responsive and clock-controlled output pathways in Lotus japonicus with special emphasis of its uniqueness. Plant Cell Physiol. 2010;51:1800–14. doi: 10.1093/pcp/pcq140. [DOI] [PubMed] [Google Scholar]
- 47.Weake V.M., Workman J.L. Inducible gene expression: diverse regulatory mechanisms. Nat. Rev. Genet. 2010;11:426–37. doi: 10.1038/nrg2781. doi:10.1038/nrg2781. [DOI] [PubMed] [Google Scholar]
- 48.Eisen M.B., Spellman P.T., Brown P.O., Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc. Natl Acad. Sci. USA. 1998;95:14863–68. doi: 10.1073/pnas.95.25.14863. doi:10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.