Abstract
Female BRCA1 mutation carriers have a nearly 80% probability of developing breast cancer during their life-time. We hypothesized that the breast epithelium at risk in BRCA1 mutation carriers harbors mammary epithelial cells (MEC) with altered proliferation and differentiation properties. Using a three-dimensional culture technique to grow MECs ex vivo, we found that the ability to form colonies, an indication of clonality, was restricted to the aldehyde dehydrogenase 1–positive fraction in MECs but not in HCC1937 BRCA1-mutant cancer cells. Primary MECs from BRCA1 mutation carriers (n = 9) had a 28% greater ability for clonal growth compared with normal controls (n= 6; P = 0.006), and their colonies were significantly larger. Colonies in controls and BRCA1 mutation carriers stained positive for BRCA1 by immunohistochemistry, and 79% of the examined single colonies from BRCA1 carriers retained heterozygosity for BRCA1 (ROH). Colonies from BRCA1 mutation carriers frequently showed high epidermal growth factor receptor (EGFR) expression (71% EGFR positive versus 44% in controls) and were negative for estrogen receptor (ERα; 32% ER negative, 44% mixed, 24% ER positive versus 90% ER positive in controls). Expression of CK14 and p63 were not significantly different. Microarray studies revealed that colonies from BRCA1-mutant PMECs anticipate expression profiles found in BRCA1-related tumors, and that the EGFR pathway is up-regulated. We conclude that BRCA1 haploin-sufficiency leads to an increased ability for clonal growth and proliferation in the PMECs of BRCA1 mutation carriers, possibly as a result of EGFR pathway activation. These altered growth and differentiation properties may render BRCA1-mutant PMECs vulnerable to transformation and predispose to the development of ER-negative, EGFR-positive breast cancers.
Introduction
Female BRCA1 mutation carriers have an 80% to 85% risk of developing breast cancer over their life-time. Given these unfavorable odds, many women opt for prophylactic mastectomies, which reduces breast cancer risk by 90% (1). Breast cancers in these patients are often of a “basaloid” phenotype, estrogen and progesteron receptor negative, histologically high-grade, epidermal growth factor (EGF) receptor (EGFR)-positive (2) and convey a poor prognosis (3). Because of the basaloid features, it has been hypothesized that these cancers are derived from early mammary epithelial cell (MEC) progenitors. In addition, the normal breast epithelium of BRCA1 mutation carriers has a high prevalence of premalignant lesions (4). These observations raise the possibility that morphologically normal PMECs with a deleterious BRCA1 mutation may have altered proliferative and differentiation patterns that predispose them to malignant transformation.
Three-dimensional basement membrane cultures provide a unique opportunity to model the architecture of epithelium in vitro (5–7). We have previously reported that PMECs from c-neu transgenic mice had a higher cloning efficiency and altered differentiation patterns when compared with wild-type mice (7). Here, we used ex vivocultures as a tool to uncover the proliferation and differentiation properties of PMECs from BRCA1 mutation carriers.
Materials and Methods
PMEC isolation
Fresh tissue was obtained according to Institutional Review Board–approved protocols, minced, and resuspended in 50 mL MEGM with Collagenase/Hyaluronidase (StemCell Technologies), incubated at 37°C with agitation at 80 to 100 rpm for 3 to 6 h, followed by centrifugation at 80 × gfor 30 s, trypsin digestion and 40 µmol/L sieve-filtration to obtain a single epithelial cell suspension.
Cell culture
HCC1937 BRCA1-mutant breast cancer cells (American Type Culture Collection) were cultured in RPMI 1640/10% fetal bovine serum; PMECs in MEGM (Lonza) supplemented with bovine pituitary extract and EGF. For three-dimensional cultures, the bottoms of 8-chamber culture slides (BD Falcon) were coated with Matrigel (BD Biosciences) and 10,000 cells were plated per well.
Immunohistochemistry
Matrigel-embedded colonies were fixed in Carnoy’s solution and paraffin-embedded. We used antibodies against Cytokeratin 14 (LL002; Neo Markers), p63 (BD Pharmingen), EGFR (clone 31G7; Zymed), BRCA1 (clone Ms110; Calbiochem), or ER-α (clone MC-20; Santa Cruz), and slides were scored blindly. The ALDEFLUOR kit (StemCell Technologies) was used according to manufacturer’s instructions using FC500 cytometer (Coulter) or sorted using a MoFlo sorter (Dako).
Morphometry and statistics
Colonies were documented using ACT-1 software connected to an Olympus SZX12 or a Nikon EclipseS100 microscope and analyzed using SIGNATURE (8). PMECs from the left or right were analyzed separately in quadruples; the two-sided t test was used to determine significance.
Microarray experiments
Ten colonies were collected and RNA was isolated using the Absolutely RNA Nanoprep kit (Stratagene). Samples were hybridized to the Human Genome U133 Plus 2.0 (Affymetrix) at the Partners Genomics Center and analyzed using the gene set comparison tool of BRB-ArrayTools 3.6 [National Cancer Institute (NCI)]. Predefined Biocarta pathways (NCI) and the BRCA1 signature in breast cancer (9) were analyzed using the functional class scoring method previously described (10).6
Loss of heterozygosity determination
DNA was extracted from either single or a pool of >5 colonies using the QIAamp Micro DNA kit (Qiagen), PCR-amplified, sequenced (3730xl DNA Analyzers; Applied Biosystems) and analyzed (Mutation Surveyor software SoftGenetics).
Results and Discussion
Aldehyde dehydrogenase 1+ MECs generate characteristic colonies in vitro
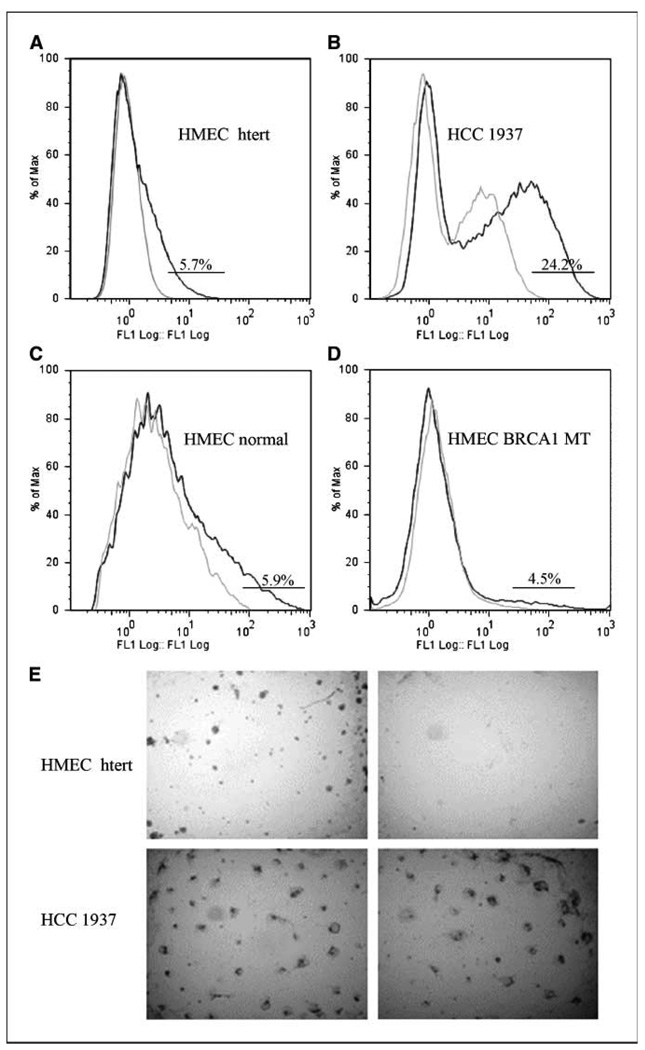
Aldehyde dehydrogenase 1 (ALDH1) has been described as a stem-cell marker for normal and malignant MECs (11). It has been reported that the normal breast epithelium of BRCA1 mutation carriers can contain acini with ALDH1-positive cells not seen in reduction mammoplasties (12). These observations suggest a crucial role for the ALDH1-positive subpopulation for the differentiation of normal MECs and for tumor formation. We used the flow cytometric ALDEFLUOR assay to identify an ALDH1-positive subpopulation in various types of MECs: HMECs immortalized by the catalytic subunit of human telomerase (HMEC htert; Fig. 1A; refs. 13, 14), normal HMECs from reduction mammoplasties (Fig. 1C), HMECs from BRCA1 mutation carriers (Fig. 1D), and breast cancer cells from the HCC1937 cell line derived from a BRCA1-deficient breast cancer (Fig. 1B; ref. 15). We found that all four types of breast epithelial cells contained an ALDH1-positive subpopulation (Fig. 1). Interestingly, HMEC htert (Fig. 1A) or freshly isolated normal or BRCA1-mutant MECs (Fig. 1C and D), all contained a 4.5% to 6% ALDH1+ population, whereas the BRCA1-mutant HCC1937 had >24% cells positive for ALDH1 (Fig. 1D). After sorting, ALDH1+ MECs proved to be highly clonal when embedded as single cells in Matrigel-based cultures, whereas the ALDH1− fraction failed to grow (Fig. 1E). Similar observations were made by Ginestier and colleagues (11), who found a frequency of 8% for ALDH1+ PMECs that gave increase to colonies in a clonogenic assay. However, in the case of HCC1937, BRCA1-mutant breast cancer cells, both the ALDH1+ and the ALDH1− fraction formed colonies with similar frequency (Fig. 1E). In summary, the breast epithelium from normal individuals and from BRCA1 mutation carriers contains ALDH1-positive MECs. These can be assessed in a three-dimensional culture assay. Clonogenic growth of HMECs is restricted to the ALDH1-positive subpopulation. Breast cancer cells, however, have a higher percentage of ALDH1+ cells, and both, the ALDH1+ and ALDH1− fractions formed colonies.
Figure 1.
The ability to form colonies ex vivo is restricted to the ALDH1-positive subpopulation in HMECs but not in breast cancer cells. ALDH1 was measured in the absence (dark line) or presence (light line) of DEAB. A, ALDH1-positive subpopulations in immortalized HMECs (HMEC htert), (B) HCC 1937 breast cancer cells, (C) HMECs isolated from a reduction mammoplasty, and (D) HMECs isolated from a BRCA1 mutation carrier. E, clonal growth in three-dimensional culture was restricted to sorted ALDH1+ in nonmalignant HMECs (top) but not in HCC1937 cancer cells (bottom).
Growth and differentiation properties of normal and BRCA1-mutant HMECs ex vivo
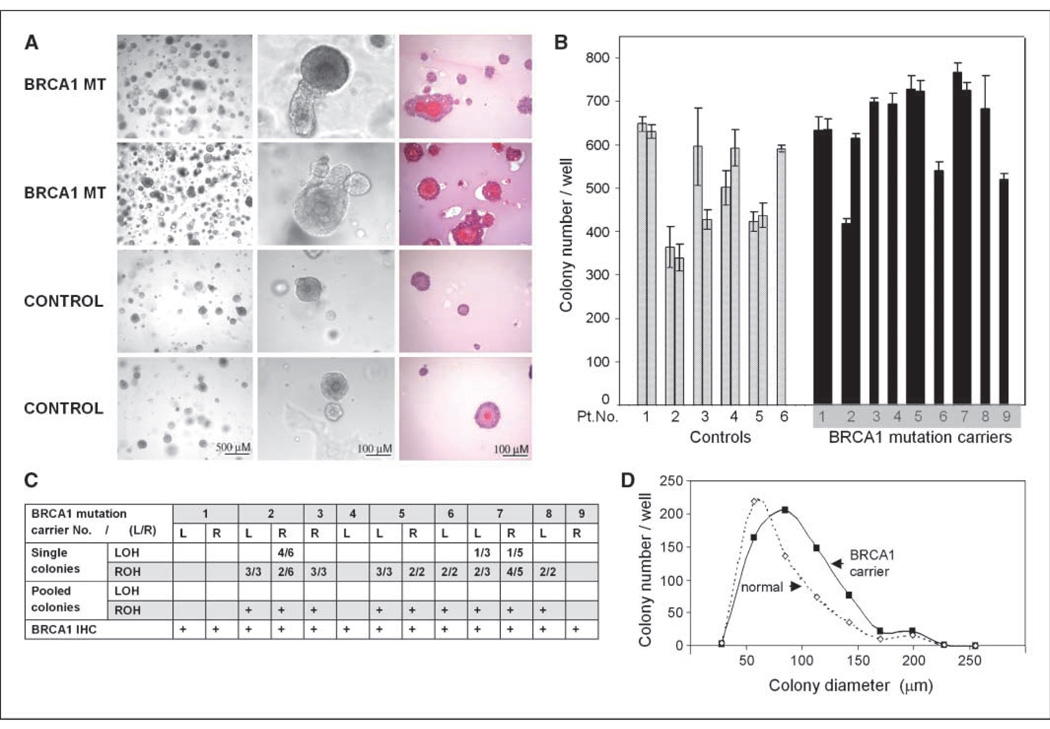
The underlying principle of colony formation assays on semisolid medium is that single progenitor cells can give rise to aggregates of terminally differentiated cells. We previously found that PMECs isolated from mice transgenic for c-neu showed disturbed phenotypes when differentiated in ex vivo cultures, long before they formed tumors in vivo (7). Here, we used this assay to determine differences between HMECs from reduction mammoplasties and from BRCA1 mutation carriers undergoing prophylactic mastectomies. Cultures succeeded in 9 of 10 of BRCA1 mutation carriers and in 6 of 12 of normals (Table 1) with colony formation efficiency ranging between 3% to 8% (Fig. 2), similar to the efficiency seen in mammosphere assays (12). Results were mostly consistent between the left and the right breast (Fig. 2). There was considerable interindividual variation in both groups (normal and BRCA1 mutation carrier), and no correlation with age. However, PMECs from BRCA1 mutation carriers had a 28% higher plating efficiency than those from normal controls (mean colony count in normals 505 versus 645 in BRCA1 mutation carriers; P = 0.006; Fig. 2B), indicative of an increased clonal growth of the BRCA1-mutant MECs. BRCA1 mutation carriers had a higher frequency of colonies that were larger than 115 µm (40%) than normal controls (27%), indicating a higher proliferative potential of their progenitor cells in addition to the increased clonality (Fig. 2D). Morphologically, the colonies derived from normal HMECs tended to be simple cystic structures with a hollow lumen, whereas colonies derived from BRCA1 mutation carriers tended to be more complex with multiple layers of cells, at times disorganized (Fig. 2). Tumors in BRCA1 mutation carriers often show loss for wild-type BRCA1 [loss of heterozygosity (LOH); ref. 16], and Liu and colleagues (12) reported that mammary stem cell expansion is also associated with LOH for BRCA1. We therefore examined the BRCA1 status in the PMEC-derived colonies and found retention of heterozygosity (ROH) in 79% of single colonies examined, and ROH in all pools of 5 to 10 colonies (Fig. 2C). Positive staining for BRCA1 (Fig. 3) confirmed the presence of the BRCA1 protein in colonies from both, normal controls and BRCA1 mutation carriers. Our data show that BRCA1 haploinsufficiency is associated with the phenotypes of increased clonality, proliferation, and altered differentiation.
Table 1.
Patient characteristics
| BRCA1 mutation | Age (y) | Ex vivo growth | Control age (y) | Ex vivo growth |
|---|---|---|---|---|
| BRCA1 185 del AG | 34 | Yes | 30 | No |
| BRCA1 185 del AG | 34 | Yes | 36 | Yes |
| BRCA1 5385 ins C | 40 | Yes | 36 | No |
| BRCA1 185 del AG | 40 | Yes | 42 | Yes |
| BRCA1 4184 del4 | 43 | Yes | 42 | Yes |
| BRCA1 185 del AG | 44 | No | 44 | No |
| BRCA1 ivs5+1G>C | 48 | Yes | 46 | No |
| BRCA1 5385 ins c | 51 | Yes | 52 | Yes |
| BRCA1 5385 ins c | 64 | Yes | 54 | Yes |
| BRCA1 Y 1522 C | 65 | Yes | 59 | No |
| 63 | No | |||
| 72 | Yes |
NOTE: Primary MECs were isolated from 10 BRCA1 mutation carriers and 12 controls. Ex vivo cultures succeeded in 9 of 10 BRCA1 mutation carriers, and 6 of 12 reduction mammoplasties when cells were seeded at 10,000 cells per well.
Figure 2.
Progenitor cell colonies from BRCA1 mutation carriers have distinct morphologies. A, isolated HMECs from BRCA1 mutation carriers (1st and 2nd row) or reduction mammoplasties (3rd and 4th row) were grown for 12 to 14 d, photographed, and subjected to H&E stain (3rd column).C, progenitor cell colonies from BRCA1 mutation carriers mostly retain heterozygosity for wild-type BRCA1 (ROH). DNA was extracted from single or pooled (5–10) colonies and sequenced. B and D, progenitor cell colonies from BRCA1 mutation carriers are more numerous and larger than normal controls. B, total colony counts are displayed. Each column represents triplicate or quadruplicate results from one breast. Adjacent columns represent results from the left and right breast from one individual. Columns are arranged from left to right in order of ascending age. D, the mean colony count for individuals from each cohort; BRCA1 mutation carriers (filled symbol) versus normal controls (open symbol) is displayed as a function of colony diameter. The mean colony number (Y axis) for each volume channel (X axis) was determined from quadruple assays from PMECs of 9 BRCA1 mutation carriers and six controls.
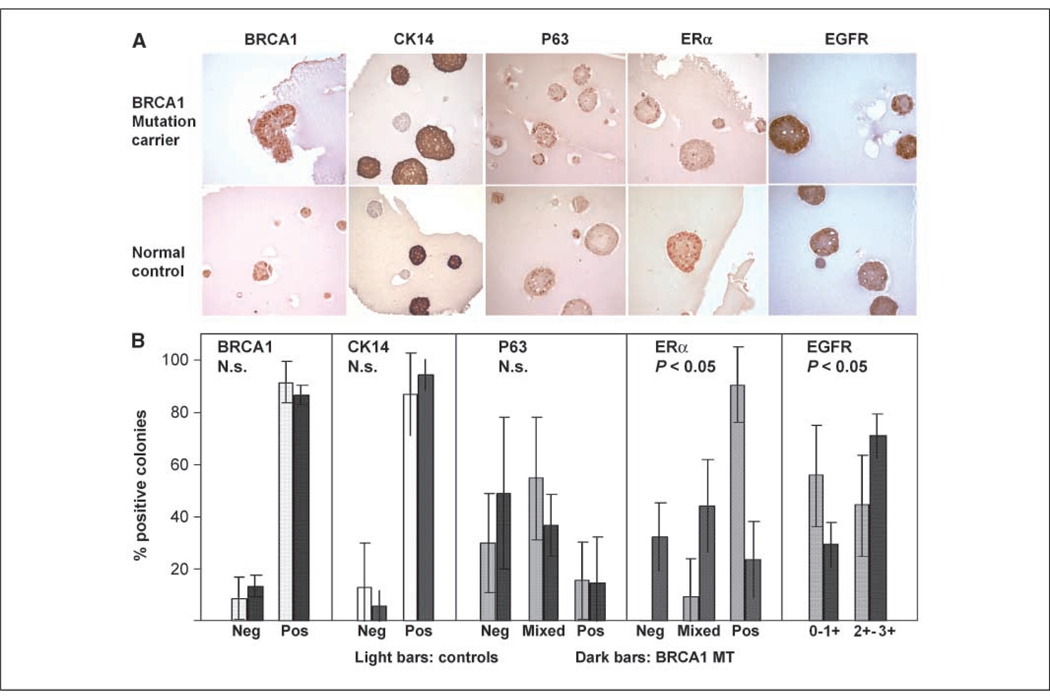
Figure 3.
Representative IHC images of colonies grown ex vivo (days 12–14). A, expression of BRCA1, lineage markers CK14 and p63, ERα, and EGFR in ex vivo colonies from BRCA1 mutation carriers (top) or normal controls (bottom). B, colonies were either positive (>80%), mixed, or negative (<10%). For EGFR, staining intensity was graded. Colonies were counted and the significance levels were calculated using a two-sided t test. Neg, negative; pos, positive; N.s., not significant.
Altered differentiation with low estrogen receptor and high EGFR expression in BRCA1-mutant PMEC colonies
In normal breast tissue, CK14 expression is restricted to the columnar basal epithelia of the ducts (17). We found that over 80% of all colonies strongly and uniformly expressed CK14 (Fig. 3), indicating that PMECs that give rise to differentiating colonies are mostly basal cells. Although their proportion was not significantly different, the absolute number of CK14-positive colonies was significantly increased in BRCA1 mutation carriers compared with normal controls (605 ± 39 versus 433 ± 80 per 10,000 cells seeded), consistent with an increased pool of basal PMECs in these patients. P63 is a marker of myoepithelial differentiation in benign and malignant breast tissue (17). There was no difference in p63 patterns between BRCA1 mutation carriers and controls, indicating that myoepithelial differentiation was not significantly different between the two groups (Fig. 3).
In normal nonlactating breast tissue, <10% of MECs express estrogen receptor (ERα; ref. 18), whereas in the lactating breast, >70% of MECs are positive for ERα (19). Because our culture conditions induced terminal differentiation, we found strong ERα expression in >90% of control colonies. In BRCA1 mutation carriers, however, ER expression was more heterogeneous with negative, mixed, and positive colonies, indicating at least a partial block toward ERα expression (Fig. 3).
Whereas EGFR expression in sporadic breast cancer tends to be <20%, high EGFR expression is seen in up to 80% of BRCA1-related breast cancer (2). Interestingly, 71% of colonies from BRCA1 mutation carriers showed strong EGFR compared with 44% of controls (Fig. 3), raising the possibility that up-regulation of EGFR is an early event in BRCA1-related tumors.
Colonies derived from BRCA1-mutant PMECs share gene expression profiles with BRCA1-related tumors and activation of the EGFR pathway
Biocarta pathway analysis revealed that BRCA1, BRCA2 and ATR, p53, and cell cycle pathways were differentially expressed between BRCA1-mutant and normal PMECs (Table 2). Furthermore, expression of genes from the BRCA1 breast cancer signature (9) that distinguished BRCA1-related from sporadic breast cancers was significantly increased in colonies derived from BRCA1-mutant PMECs, i.e., ex vivo cultures induced expression profiles subsequently found in BRCA1-related breast cancers (Table 2). Consistent with our immunohistochemistry (IHC) data (Fig. 3), we found activation of the EGFR pathway in PMEC colonies from BRCA1 mutation carriers using three different EGFR pathway data sets (Table 2).
Table 2.
Selected genes and pathways that are differentially expressed between BRCA1-mutated and normal PMECs
| Pathway | Pathway P* | Gene | Gene P† |
|---|---|---|---|
| Role of BRCA1, BRCA2 and ATR in cancer susceptibility | <0.03 | RAD51 | 0.01 |
| BRCA1 | 0.03 | ||
| MRE11A | 0.03 | ||
| CHEK1 | 0.05 | ||
| p53 signaling pathway | <0.02 | GADD45A | 0.02 |
| RB1 | 0.04 | ||
| CDKN1A | 0.05 | ||
| PCNA | 0.05 | ||
| Cell cycle | <0.00001 | SMAD2 | 0.009 |
| CCNE2 | 0.01 | ||
| CDKN2D | 0.03 | ||
| TGFB3 | 0.03 | ||
| BRCA1 signature in breast cancer | <0.02 | GPC1 | 0.009 |
| PARVA | 0.03 | ||
| CRYM | 0.04 | ||
| DUSP13 | 0.04 | ||
| EGF signaling pathway | <0.01 | AXL | 0.03 |
| JUN | 0.001 | ||
| DDIT3 | 0.04 | ||
| Role of EGFR transactivation by G-protein–coupled receptors | <0.01 | JAK1 | 0.04 |
| IGFBP2 | 0.09 | ||
| PRKCA | 0.04 | ||
| EGFR signaling pathway | <0.01 | SHC1 | 0.1 |
| EGFR | 0.1 | ||
| MDM2 | 0.1 |
Abbreviations: JAK, Janus-activated kinase; PCNA, proliferating cell nuclear antigen.
By Functional Class Scoring Method.
By t test.
In summary, we report that the nonmalignant HMECs from BRCA1 mutation carriers contain a subpopulation of progenitor cells with significantly increased clonal and proliferative potential compared with normal controls. Seventy-nine percent of these cells have not undergone LOH but are heterozygote for BRCA1 (ROH), and express BRCA1. These progenitor cells are a heterogenous population of cells that are partially blocked from differentiation to ER+ cells and have high EGFR expression. They have gene expression profiles that are subsequently found in BRCA1-related breast cancers and show activation of the EGFR-pathway. Based on these observations, we propose a model where a deleterious germline BRCA1 mutation leads to an increase in EGFR pathway activation with an increased ability for clonal growth and proliferation of mammary epithelial progenitor cells and to altered differentiation patterns. These phenotypes precede LOH and may render these progenitor cells vulnerable to cancerous transformation and the development of ER-negative tumors.
Acknowledgments
Grant support: Susan Komen Foundation (BCTR0601030), Alternatives Research and Development Foundation, Specialized Programs of Research Excellence CA-089393 (NIH), and K08-CA093655 (NIH; G. Wulf).
We thank the women who donated their tissues for these studies, Dr. Christina Gewinner for donating the HMEC htert cells, and Drs. Lowell Schnipper, Steven Come, Stuart Schnitt, Hannah Gilmore, Kara Coffey, and Hallie Kasper (BIDMC) for their support
Footnotes
Reprints and Subscriptions To order reprints of this article or to subscribe to the journal, contact the AACR Publications Department at pubs@aacr.org.
http://www.ncbi.nlm.nih.gov/geo; accession number GSE13671.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Rebbeck TR, Friebel T, Lynch HT, et al. Bilateral prophylactic mastectomy reduces breast cancer risk in BRCA1 and BRCA2 mutation carriers: the PROSE Study Group. J Clin Oncol. 2004;22:1055–1062. doi: 10.1200/JCO.2004.04.188. [DOI] [PubMed] [Google Scholar]
- 2.van der Groep P, Bouter A, van der Zanden R, et al. Distinction between hereditary and sporadic breast cancer on the basis of clinicopathological data. J Clin Pathol. 2006;59:611–617. doi: 10.1136/jcp.2005.032151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van de Rijn M, Perou CM, Tibshirani R, et al. Expression of cytokeratins 17 and 5 identifies a group of breast carcinomas with poor clinical outcome. Am J Pathol. 2002;161:1991–1996. doi: 10.1016/S0002-9440(10)64476-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hoogerbrugge N, Bult P, Bonenkamp JJ, et al. Numerous high-risk epithelial lesions in familial breast cancer. Eur J Cancer. 2006;42:2492–2498. doi: 10.1016/j.ejca.2006.05.027. [DOI] [PubMed] [Google Scholar]
- 5.Barcellos-Hoff MH, Aggeler J, Ram TG, Bissell MJ. Functional differentiation and alveolar morphogenesis of primary mammary cultures on reconstituted basement membrane. Development. 1989;105:223–235. doi: 10.1242/dev.105.2.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Debnath J, Muthuswamy SK, Brugge JS. Morphogenesis and oncogenesis of MCF-10A mammary epithelial acini grown in three-dimensional basement membrane cultures. Methods. 2003;30:256–268. doi: 10.1016/s1046-2023(03)00032-x. [DOI] [PubMed] [Google Scholar]
- 7.Wulf G, Garg P, Liou YC, Iglehart D, Lu KP. Modeling breast cancer in vivo and ex vivo reveals an essential role of Pin1 in tumorigenesis. Embo J. 2004;23:3397–3407. doi: 10.1038/sj.emboj.7600323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen JZ, Grigorieff N. SIGNATURE: a single-particle selection system for molecular electron microscopy. J Struct Biol. 2007;157:168–173. doi: 10.1016/j.jsb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 9.van ’t Veer LJ, Dai H, van de Vijver MJ, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. doi: 10.1038/415530a. [DOI] [PubMed] [Google Scholar]
- 10.Pavlidis P, Qin J, Arango V, Mann JJ, Sibille E. Using the gene ontology for microarray data mining: a comparison of methods and application to age effects in human prefrontal cortex. Neurochem Res. 2004;29:1213–1222. doi: 10.1023/b:nere.0000023608.29741.45. [DOI] [PubMed] [Google Scholar]
- 11.Ginestier C, Hur MH, Charafe-Jauffret E, et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell. 2007;1:555–567. doi: 10.1016/j.stem.2007.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu S, Ginestier C, Charafe-Jauffret E, et al. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A. 2008;105:1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang XR, Jimenez G, Chang E, et al. Telomerase expression in human somatic cells does not induce changes associated with a transformed phenotype. Nat Genet. 1999;21:111–114. doi: 10.1038/5056. [DOI] [PubMed] [Google Scholar]
- 14.Stampfer MR, Garbe J, Levine G, Lichtsteiner S, Vasserot AP, Yaswen P. Expression of the telomerase catalytic subunit, hTERT, induces resistance to transforming growth factor β growth inhibition in p16INK4A(−) human mammary epithelial cells. Proc Nad Acad Sci U S A. 2001;98:4498–4503. doi: 10.1073/pnas.071483998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Scully R, Ganesan S, Vlasakova K, Chen J, Socolovsky M, Livingston DM. Genetic analysis of BRCA1 function in a defined tumor cell line. Mol Cell. 1999;4:1093–1099. doi: 10.1016/s1097-2765(00)80238-5. [DOI] [PubMed] [Google Scholar]
- 16.Smith SA, Easton DF, Evans DG, Ponder BA. Allele losses in the region 17q12–21 in familial breast and ovarian cancer involve the wild-type chromosome. Nat Genet. 1992;2:128–131. doi: 10.1038/ng1092-128. [DOI] [PubMed] [Google Scholar]
- 17.Li H, Cherukuri P, Li N, et al. Nestin is expressed in the basal/myoepithelial layer of the mammary gland and is a selective marker of basal epithelial breast tumors. Cancer Res. 2007;67:501–510. doi: 10.1158/0008-5472.CAN-05-4571. [DOI] [PubMed] [Google Scholar]
- 18.Petersen OW, Hoyer PE, van Deurs B. Frequency and distribution of estrogen receptor-positive cells in normal, nonlactating human breast tissue. Cancer Res. 1987;47:5748–5751. [PubMed] [Google Scholar]
- 19.Saji S, Jensen EV, Nilsson S, Rylander T, Warner M, Gustafsson JA. Estrogen receptors α and β in the rodent mammary gland. Proc Natl Acad Sci U S A. 2000;97:337–342. doi: 10.1073/pnas.97.1.337. [DOI] [PMC free article] [PubMed] [Google Scholar]