Summary
It has been proposed that an aggressive secondary cancer stem cell population arises from a primary cancer stem cell population through acquisition of additional genetic mutations and drives cancer progression. Overexpression of Polycomb protein EZH2, essential in stem cell self-renewal, has been linked to breast cancer progression. However, critical mechanism linking increased EZH2 expression to BTIC (breast tumor initiating cell) regulation and cancer progression remains unclear. Here, we identify a mechanism in which EZH2 expression-mediated downregulation of DNA damage repair leads to accumulation of recurrent RAF1 gene amplification in BTICs, which activates p-ERK-β-catenin signaling to promote BTIC expansion. We further reveal that AZD6244, a clinical trial drug that inhibits RAF1-ERK signaling, could prevent breast cancer progression by eliminating BTICs.
Introduction
Accumulated evidence shows that cancer stem/progenitor cells (or tumor initiating cells, TICs) account for cancer initiation, progression and drug resistance (Al-Hajj and Clarke, 2004; Reya et al., 2001; Rossi et al., 2008). TICs in different types of cancers have been gradually identified, for example, breast tumor initiating cells (BTICs) can be isolated by sorting for CD44+CD24−/low cells (Al-Hajj et al., 2003) or Hoechst negative side population (SP) cells (Patrawala et al., 2005), and can also be enriched by suspension in spheroid culture termed mammospheres and serial transplantations in immunodeficient mice (Ponti et al., 2005). Compared to differentiated cells/non-stem cells with limited opportunity to accumulate multiple lesions, long-lived stem/progenitor cells allow progression towards malignancy through accumulation of epigenetic or genetic alterations that deregulate self-renewal pathways (Rossi et al., 2008). It was proposed that a more aggressive secondary cancer stem/progenitor cell population may arise from a primary cancer stem cell population through acquisition of additional genetic mutations that deregulate cancer stem/progenitor homeostasis and drive cancer progression (Visvader and Lindeman, 2008). Moreover, studies showed that high grade tumors are enriched with a high content of TICs (Pece et al.). However, the key components and molecular mechanisms contributing to TIC formation or expansion are largely unknown.
Epigenetic regulation by Polycomb proteins is essential in maintaining the self-renewal capability of embryonic and adult stem cells through mediating histone methylation at lysine 27 of histone H3 (H3K27) (Cao and Zhang, 2004; Ezhkova et al., 2009; Lessard and Sauvageau, 2003; Sparmann and van Lohuizen, 2006). Interestingly, high expression of EZH2, a key component of the Polycomb PRC2 complex, has been linked to aggressive progression of breast and prostate cancers (Kleer et al., 2003; Varambally et al., 2002). However, critical mechanisms linking increased EZH2 expression to BTIC regulation and cancer progression remain unclear.
The importance of the tumor microenvironment in cancer has been increasingly recognized (Hu and Polyak, 2008). The microenvironment of solid tumors contains regions of poor oxygenation as a result of hypoxia. It is worthy to note that hypoxia/HIF1α activation is associated with high grade basal breast cancer and poor prognosis (Bristow and Hill, 2008). Using microarray-based gene expression profiles, previous studies have identified a group of DNA damage repair genes that are downregulated by hypoxia, including RAD51 (Bindra et al., 2007; Bindra and Glazer, 2007; Bindra et al., 2004). Interestingly, forced expression of EZH2 in breast epithelial cells correlates with decreased expression of double-strand-break repair protein RAD51 paralogs through an unknown mechanism (Zeidler et al., 2005). Whether hypoxia associates with EZH2 on regulation of DNA damage repair remains to be explored. More importantly, disruption of critical DNA damage repair proteins, such as RAD51, is expected to result in a significant increase of spontaneous chromosomal break and chromosome instability (Dodson et al., 2004), which could further lead to oncogenic translocation and amplification (Difilippantonio et al., 2002).
This study is to identify a mechanism linking increased EZH2 expression to BTIC regulation and cancer progression.
Results
Increased EZH2 expression in BTICs is linked to decreased RAD51 expression, enhanced BTICs and high grade breast cancer
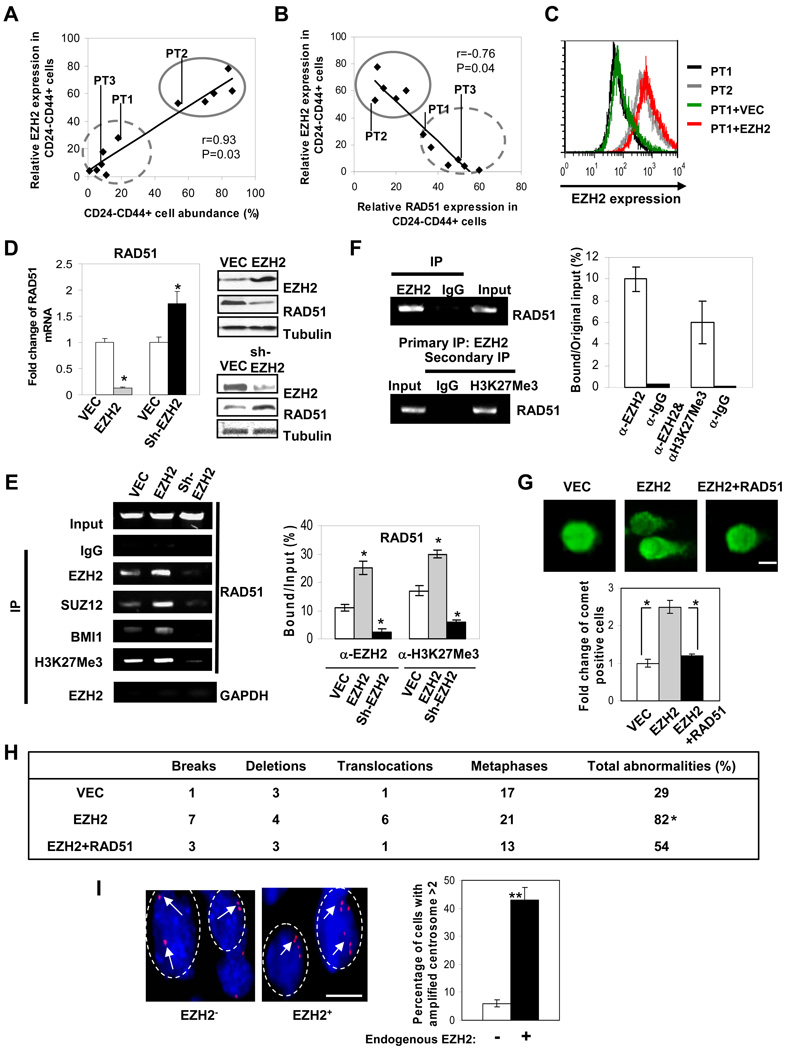
To understand whether EZH2 expression is involved in regulation of BTICs and cancer progression, we examined the endogenous EZH2 expression level in the CD44+CD24−/low cells isolated from human breast cancer and non-cancer cell lines, xenograft tumor cells, and primary breast tumor cells. We found that EZH2 levels in the CD44+CD24−/low cells positively correlate with the percentage of CD44+CD24−/low cells (Figure 1A). EZH2 expression was also enriched in the primary tumor cell populations highly expressing ALDH1 (ALDH+) and OCT4 (OCT4+) (Figure S1A), both of which are known markers of cancer stem cells/stem cells (Wicha, 2008). These data suggest EZH2 expression level may play a role in regulating the BTIC population.
Figure 1. Elevated expression of EZH2 in BTICs reduces RAD51 expression and increases genomic abnormality.
(A, B) Intracellular staining correlating EZH2 expression with the percentage of CD44+CD24−/low cell population (A) or with RAD51 expression in CD44+CD24−/low cells from high grade tumor cells (B) using flow cytometry. Solid circle: Patient Tumor-PT2, basal-like breast cancer cells-MDA-MB-231, Hs578t, BT549, BT549 xenograft tumor cells; dotted circle: Patient Tumor-PT1, Patient Tumor-PT3, low grade breast cancer cell lines-SKBR3, MCF7, MCF7 xenograft tumor cells, and benign mammary cells MCF10A. (C) Intracellular staining shows endogenous EZH2 and ectopically expressing EZH2 levels in CD44+CD24−/low population isolated from human primary tumor cells. Infected human primary tumor cells in suspension culture enriched for CD44+CD24−/low were subjected to (D) realtime PCR and immunoblotting; (E)(F) chromatin immunoprecipitation (ChIP, PCR product encompassing GAGA motif); (G) comet analysis shown as representative fluorescence microscopy images (scale bar: 5µm). (H) Table summarizing the percentage of CD44+CD24−/low enriched-human primary tumor cells exhibiting chromosome aberrations per metaphase by karyotype analysis (n=6, duplicate in each group). (I) Percentage of the cells with amplified red-CEP3 centrosomes with high or low EZH2 expression (8 breast tumor sections/group, 50 nuclei were counted in each section; n>2 per nucleus is considered as amplification, scale bar: 5µm). Endogenous EZH2 expression was determined by immunohistochemical staining from Figure 4G. (−): negative-low, (+): positive-high. Error bars denote ±SD (*P<0.05, **P<0.01). See also Figure S1.
We then investigated whether EZH2 enables the accumulation of genomic/genetic abnormalities to promote BTICs through repressing RAD51. Thus, similar to Figure 1A, we examined the endogenous RAD51 expression levels in the CD44+CD24−/low cells isolated from the abovementioned samples. We found that EZH2 expression was negatively correlated with RAD51 expression in the CD44+CD24−/low cells (Figure 1B). Furthermore, EZH2 expression was most elevated whereas RAD51 expression was most repressed in the CD44+CD24−/low cells isolated from high grade/basal-like breast cancer cells (Figure 1A and 1B), which are known to be associated with aggressive malignancy, high tumor grade and poor prognosis, compared to those from less invasive/low tumor grade breast cancer cells and to those from benign mammary cell line (Figure 1A and 1B). Consistent with the primary cells and cell line data, high EZH2 expression was also significantly correlated with high tumor grade in a cohort of human breast tumor sections (Table 1), where EZH2 expression was negatively correlated with RAD51 expression (Table 2). Together, these data suggest a link between EZH2 and RAD51 expression in the regulation of BTICs and cancer progression.
Table 1. EZH2 expression is positively correlated with tumor grade.
Expression patterns of EZH2 and tumor grade in the consecutive sections from 128 human breast tumors were determined and summarized. (−): negative-low, (+): positive-high. The correlation between EZH2 and tumor grade was analyzed using SPSS 16.0 Pearson Chi-Square test.
| Tumor Grade | I | II&III | Total |
|---|---|---|---|
| EZH2 negative/low | 13 (10.2%) | 24 (18.7%) | 37 (28.9%) |
| EZH2 high | 13 (10.2%) | 78 (60.9%) | 91 (71.1%) |
| Total | 26 (20.4%) | 102 (79.6%) | 128 (100%) |
| P<0.008 |
Table 2. EZH2 expression negatively correlates with RAD51 but positively with RAF1, p-ERK and β-catenin expression in breast carcinoma.
Expression patterns of EZH2 and RAD51; EZH2 and RAF1; EZH2 and p-ERK; EZH2 and β-catenin in the consecutive sections from human breast tumors were determined and summarized. (−): negative-low, (+): positive-high. The correlation between EZH2, RAF1, RAD51, p-ERK and β-catenin was analyzed using SPSS 16.0 Pearson Chi-Square test.
| EZH2 | RAD51 | RAF1 | p-ERK | β-catenin (C+N) | ||||
|---|---|---|---|---|---|---|---|---|
| − | + | − | + | − | + | − | + | |
| − | 11(17.5%) | 26(41.3%) | 61(49%) | 2(2%) | 35(28.2%) | 28(22.6%) | 40(38.1%) | 27(25.7%) |
| + | 14(22.2%) | 12(19.0%) | 25(20%) | 36(29%) | 19(15.3%) | 42(33.9%) | 8(7.6%) | 30(28.6%) |
| Total | 26(39.7%) | 38(60.3%) | 86(69%) | 38(31%) | 54(43.5%) | 70(56.5%) | 48(45.7%) | 57(54.3%) |
| P=0.003 | P<0.001 | P=0.013 | P=0.001 | |||||
Elevated expression of EZH2 in BTICs downregulates RAD51 expression and increases genomic abnormality
To investigate if elevated EZH2 expression indeed plays a role in promoting BTICs, human primary tumor cells enriched for BTICs by sorting with CD44+CD24−/low marker were infected with retrovirus expressing EZH2 or other indicated constructs (Figure S1B), and then maintained in suspension culture to produce mammospheres. Mammospheres still contained 87% CD44+CD24−/low cells after 7 days of culture in suspension and could be differentiated in conditions as previously described (Dontu et al., 2003) and (Ponti et al., 2005) (Figure S1C). To ensure EZH2 expression level is physiological relevant, we purposely increased the EZH2 level in the low tumor grade BTICs (PT1) to a level that is comparable to the physiologically high EZH2 level in the high tumor grade BTICs (PT2) (PT1+EZH2 vs. PT2, Figure 1C). We found that ectopic expression of EZH2 downregulated RAD51 mRNA and protein in the BTICs from human primary tumor cells (Figure 1D, left and upper right), whereas knocking-down EZH2 using small hairpin RNA (shRNA) increased RAD51 mRNA and protein expression (Figure 1D, left and lower right). Through chromatin immunoprecipitation (ChIP), we found that the PRC2 complex proteins including EZH2 and SUZ12 as well as PRC1 component (BMI1) (Figure 1E) were recruited and bound to the putative Polycomb response element (PRE) containing PHO motif and GAGA motif within the RAD51 promoter (Figure S1D) (Sing et al., 2009), where EZH2 and H3K27Me3 co-occupied (Figure 1F). In addition, RAD51 transcription was suppressed by wild-type EZH2 but not by the methyl-transferase deleted mutant (ΔSET), suggesting that H3K27 histone methylation mediated by EZH2 involves in the silencing of RAD51 transcription (Figure S1E). Accordingly, downregulation of EZH2 by shRNA reduced H3K27 histone methylation within RAD51 promoter (Figure 1E), which could thereby increase RAD51 expression as shown in Figure 1D.
To determine the biological effects resulting from EZH2-RAD51 regulation, we examined changes in DNA lesions/aberrations given the critical role of RAD51 in DNA damage repair. We found that EZH2-mediated downregulation of RAD51 induced a substantial amount of spontaneous DNA damage indicated by the increased comet tail formation (Singh et al., 1988), which was blocked by co-expression of a retroviral promoter driven-RAD51 plasmid that is free from the transcriptional repression by EZH2 (Figure 1G). Furthermore, ectopic EZH2 expression in BTIC-enriched cells significantly increased chromosome karyotype aberrations, including chromosome breaks, deletions and translocations, which were reduced by co-expression of RAD51 (Figure 1H, representative morphology of abnormal karyotypes is shown in Figure S1F). Next we inquired whether physiological expression level of EZH2 is also related to chromosomal abnormalities in a cohort of tumor samples. It is noted that centrosome amplification, a hallmark of genomic instability, has been observed in cells deficient in DNA damage repair proteins, such as RAD51 (Dodson et al., 2004). Thus, we examined endogenous EZH2 level and centrosome number in a cohort of human breast tumors. We found that high expression level of endogenous EZH2 significantly correlates with high centrosome amplification (Figure 1I), indicating the clinical relevance of EZH2-enhanced genomic instability in cancer. Taken together, EZH2 likely plays a role in accumulating genomic abnormalities through repression of RAD51 expression in the BTIC-enriched population, which may lead to deregulated signaling required for tumor progression.
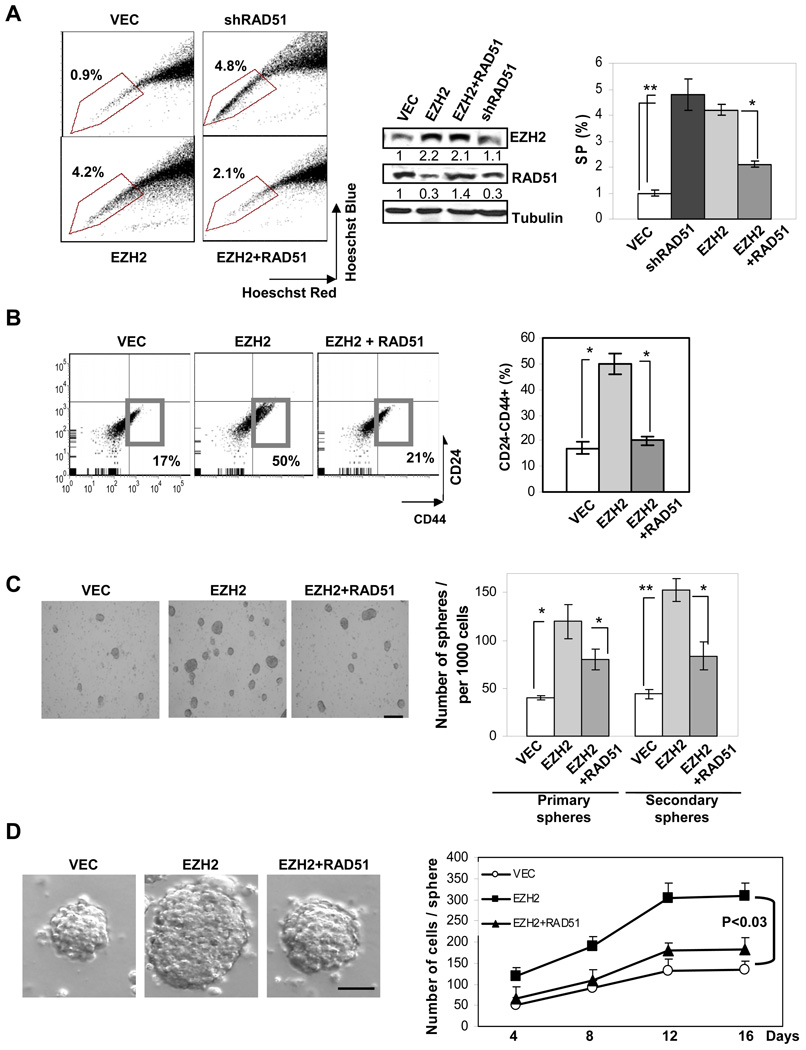
Increased EZH2 expression enhances self-renewal and expansion of BTICs
Next, we asked whether EZH2-RAD51 regulation directly affects BTICs. We found that ectopically expressing EZH2 in human primary breast tumor cells increased the percentage of SP cells (Figure 2A) and CD44+CD24−/low cells (Figure 2B), which was suppressed upon RAD51 co-expression (Figure 2A and Figure 2B). Similar to ectopically expressing EZH2, knocking down RAD51 significantly enhanced the abundance of SP cells (Figure 2A). To determine if EZH2 expands BTICs through enhancing the self-renewing capacity, we examined serially propagated mammosphere formation in vitro and mammary xenograft tumor formation in vivo using primary tumor cells (PT1). Increased expression of EZH2 effectively enhanced primary and secondary mammosphere formation (Figure 2C), which could generate xenograft tumors with 10-fold fewer cells than the control group (Please see later, Table S3). Moreover, the mammospheres expressing EZH2 contained 2–3 fold more cells per sphere, leading to enlarged sphere size (Figure 2D), and suggesting EZH2 may also promote BTIC proliferation and/or survival. In contrast, RAD51 co-expression, at least in part, dampened the EZH2 enhanced mammosphere number and size (Figure 2C–2D). Similar results were observed in BTIC-enriched sphere cultures generated from two other primary tumor samples (data not shown). Taken together, our data suggest that EZH2 expands highly self-renewing BTICs through the downregulation of RAD51.
Figure 2. EZH2 enhances self-renewal and expansion of BTICs.
Infected human primary breast tumor cells in suspension culture were used to determine the percentage of (A) SP (marked by polygon) and (B) CD44+CD24−/low (marked by square) populations by flow cytometry. (C) The number of primary and secondary mammospheres expressing the indicated constructs was counted per 1000 cells (scale bar: 200µm). (D) Cell number was counted from dissociated human primary mammospheres collected at the indicated time point (scale bar: 100µm). Error bars denote ±SD (*P<0.05, **P<0.01).
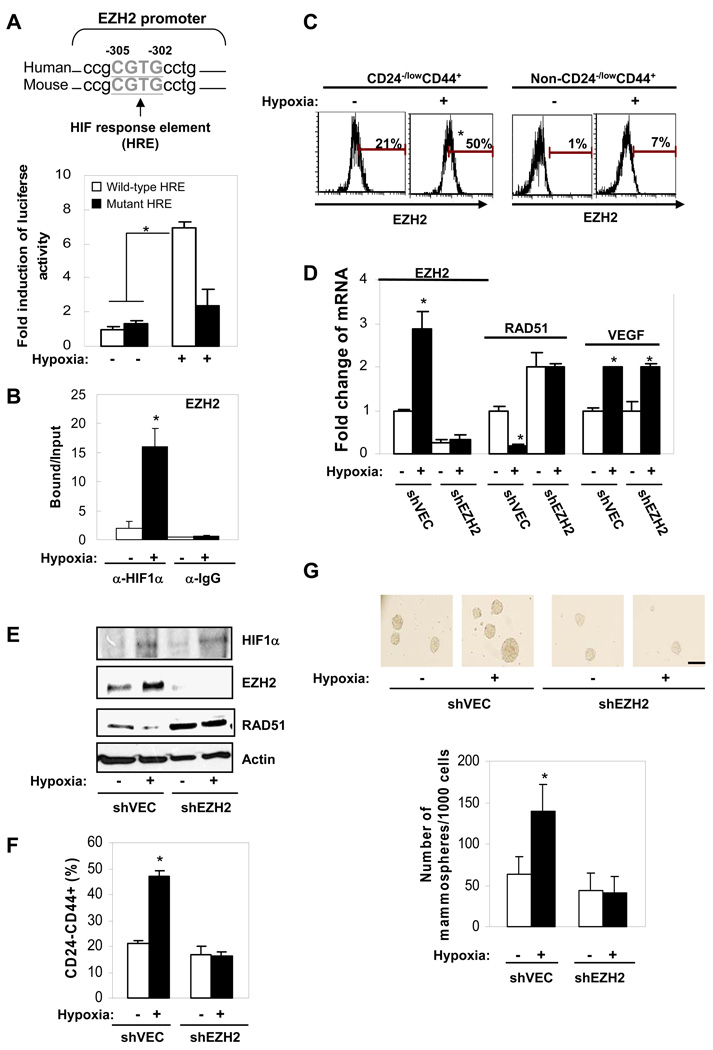
Hypoxia physiologically increases EZH2 expression, decreases RAD51 transcription and enhances BTICs
To further investigate the physiological relevance of EZH2 expression promotion of BTICs, we looked for physiological relevant conditions that may be involved in the regulation of EZH2 expression. Using promoter analysis (Genomatix®), we found a consensus sequence of HIF response element (HRE: NCGTG) within EZH2 promoter region. To test whether hypoxia might regulate EZH2 expression, we subjected primary breast tumor cells (PT1) to normoxic (20% O2) and hypoxic (1% O2) conditions and found that hypoxia significantly induces luciferase activity driven by EZH2 promoter containing a wild-type HRE, but not driven by EZH2 promoter with a mutated HRE (Figure 3A). Mutation of the HRE abolished induction of EZH2 luciferase activity by hypoxia, suggesting HIF transcription factor may mediate EZH2 activation by hypoxia (Figure 3A). Indeed, using a specific HIF1α antibody for ChIP analysis, we found HIF1α directly binds to the HRE-containing EZH2 promoter under hypoxic conditions (Figure 3B). These data suggest that upregulated HIF1α upon hypoxia treatment likely transactivates EZH2 gene and increases EZH2 expression. We next asked whether hypoxia may induce EZH2 expression in BTICs and found that the high level of EZH2 was further increased by hypoxia in CD44+CD24−/low BTICs, however, the EZH2 level remained low in non-CD44+CD24−/low cells even under hypoxia conditions (Figure 3C). In support of the results in Figure 2, RAD51 mRNA (Figure 3D) and protein (Figure 3E) levels were significantly repressed by increased EZH2 expression under hypoxia in BTIC-enriched cells, and knocking-down EZH2 by shRNA reversed hypoxia mediated-RAD51 repression (Figure 3D–3E). Knocking-down EZH2 also abolished the induction of BTICs (Figure 3F) and mammosphere formation (Figure 3G) by hypoxia, suggesting that hypoxia-mediated EZH2 upregulation was required for both the increased hypoxia-enhanced CD44+CD24−/low cell population and mammosphere formation. Together, our results suggest EZH2 expression is induced by a hypoxic microenvironment, leading to repression of RAD51 (Figure 3D–3E) and enhancement of BTIC population (Figure 3F–3G). These data provide additional support for the physiological link between induction of EZH2 expression, repression of RAD51 and enhancement of BTIC population, which are consistent with the results obtained by using the ectopic expression system.
Figure 3. Hypoxia physiologically increases EZH2 expression, decreases RAD51 transcription and enhances abundance of CD44+CD24−/low BTICs.
Primary tumor cells were treated with normoxia (20%O2) or hypoxia (1%O2) for 48 hrs and cultured in suspension for 3 days. (A) Bar graph showing fold change of EZH2 luciferase reporter analysis. The sequence alignment of HIF response elements within EZH2 promoter is shown below. (B) Bar graph showing HIF1α binds to EZH2 promoter using chromatin immunoprecipitation. (C) Histogram plots showing EZH2 expression in CD44+CD24−/low enriched-human primary tumor cells using surface and intracellular staining. Bars showing percentage of the indicated protein expression after being normalized to isotype control. (D) Fold change of mRNA expression using real time PCR in CD44+CD24−/low-enriched human primary tumor cells. VEGF was served as a positive control target gene for HIF1α. (E) Expression change of the proteins in CD44+CD24−/low-enriched-population was examined using immunoblotting. (F) The percentage of CD44+CD24−/low population in hypoxia treated-human primary tumor cell was determined using flow cytometry. (G) The number of mammospheres counted per 1000 cells (scale bar: 100µm). Error bars denote ±SD (*P<0.05).
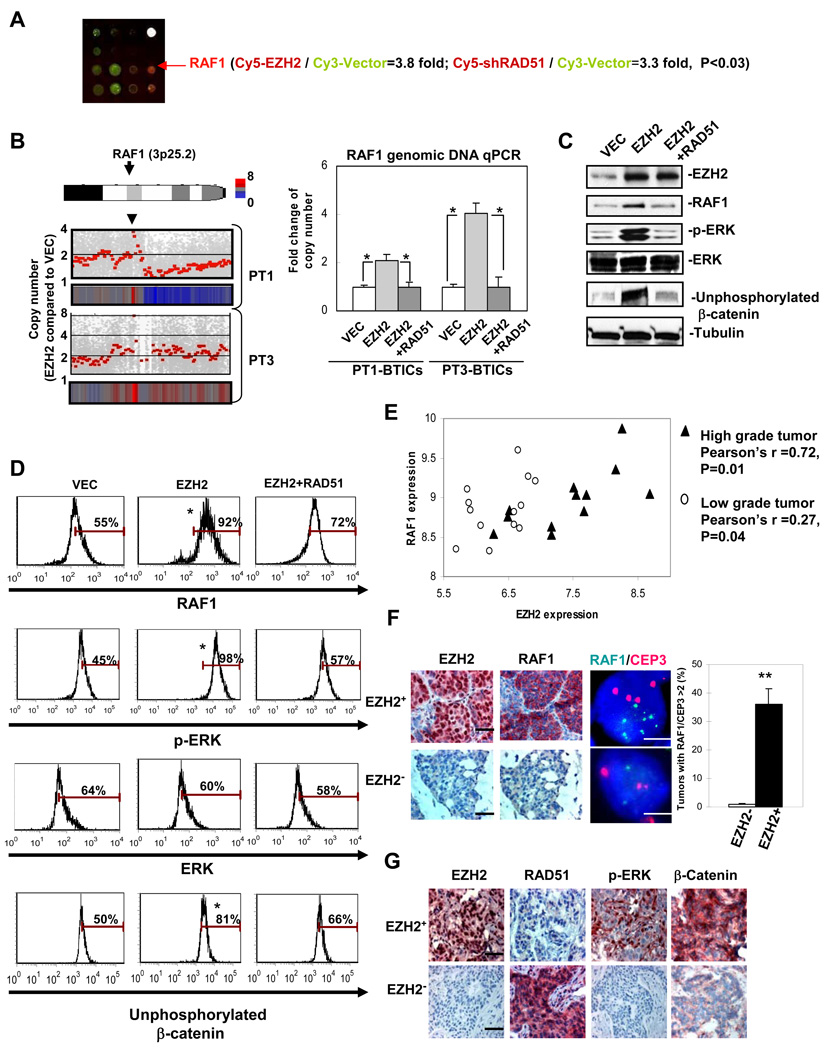
EZH2-RAD51 regulation induces RAF1 amplification and downstream p-ERK-β-catenin activation in BTICs
To further characterize the downstream target(s) that mediate BTIC promotion by EZH2-RAD51 regulation, we cross referenced the results of two protein arrays generated with lysates from BTIC enriched-cells expressing EZH2 and shRAD51. Compared to the vector control, ectopically expressing EZH2 or knocking-down RAD51 in BTIC enriched-cells most significantly increased RAF1 protein expression (Figure 4A, Table. S1). Since downregulation of RAD51 induces double strand breaks and chromosome abnormalities, such as gene amplification, we examined if the enhanced RAF1 expression is due to amplification of the RAF1 gene. Using an unbiased SNP array analysis to examine genomic copy number variation in BTIC enriched-cells expressing vector control compared with BTIC enriched-cells expressing EZH2, we found that, indeed, RAF1 gene copy number was amplified by high EZH2 expression (Figure 4B left panel, Table S2), which was further validated by quantitative PCR analysis using genomic DNA of BTIC enriched-cells from two human primary tumor cells, PT1 and PT3 (Figure 4B right panel).
Figure 4. EZH2-RAD51 regulation induces RAF1 amplification and downstream p-ERK-β-catenin activation, correlating with aggravated clinical malignancy.
(A) Schematic diagram showing protein array analysis using Panorama (Sigma) antibody arrays with lysates of the CD44+CD24−/low BTIC enriched-human primary tumor cells expressing control vector (labeled with Cy3) and EZH2/shRAD51 (labeled with Cy5, n=4/group). (B) Diagram showing copy number variation analysis (EZH2/VEC) using SNP array analysis. Results from BTIC enriched-human primary tumor samples (PT1 and PT3) are presented (left, n=3/tumor). All vector control samples were examined against two HapMap normal data sets (Affymetrix) and no RAF1 amplification was observed in any of vector controls. Quantitative genomic DNA copy number analysis was performed in these BTIC enriched-human primary tumor cells using specific probe against RAF1 gene (right). Samples were collected at 96hrs after EZH2 expression. Protein expression change in sorted CD44+CD24−/low cells from PT1 was examined using immunoblotting (C) and intracellular staining (D) using flow cytometry. Bars showing percentage of the indicated protein expression after being normalized to isotype control. (E) Correlation of EZH2 and RAF1 mRNA expression from a cohort of low grade (stage1) and high grade (stage 3–4) human breast tumors as described (Chin et al., 2006). Pearson correlation coefficient shows significant correlation r>0.7 between expression of RAF1 and EZH2 in high grade tumors. (F) A representative case exhibiting breast tumor sections analyzed by immunohistochemical staining (black scale bar: 20µm) and FISH (white scale bar: 5µm) for RAF1 amplification (green) and centromere control (CEP3, red) (corresponding to EZH2 status in Table1, n=128). Signal ratio green/red was counted in 50 nuclei from each section. Bar graph showing percentage of the counted nuclei with RAF1 amplification (signal ratio green/red>2). (G) 124 surgical specimens from breast cancer patients were analyzed by immunohistochemical staining (scale bar: 50µm). Consecutive sections from a representative case were stained using the indicated antibodies. The staining pattern (−): negative-low, (+): positive-high, (N): nuclear staining, and (C): cytoplasmic staining. Error bars denote ±SD (*P<0.05, **P<0.01). See also Figure S2, Table S1 and Table S2.
Previously we showed that activated ERK, a prime target downstream of RAF1, enhances stabilization of functional β-catenin in the nucleus through inhibiting GSK3β-meidated phosphorylation of N-terminus of β-catenin (Ding et al., 2005). It is worth noting that unphosphorylated (activated) β-catenin plays important roles in maintaining TIC survival, proliferation and self renewal (Guo et al., 2008; Li et al., 2003; Reya et al., 2003; Taipale and Beachy, 2001). Increased unphosphorylated (activated) β-catenin also contributes to radiation-resistance in BTICs (Chen et al., 2007). To understand whether EZH2 induced-RAF1 amplification activates the ERK-β-catenin signaling pathway in BTICs, we examined expression of RAF1, p-ERK and unphosphorylated β-catenin by immunoblotting and intracellular staining using an established antibody that only recognizes unphosphorylated (activated) β-catenin (Guo et al., 2008; Chen et al., 2007). Indeed, we found that RAF1 amplification is concurrently correlated with the upregulation of RAF1 downstream targets p-ERK and unphosphorylated β-catenin in CD44+CD24−/low BTICs isolated from human primary tumor cells (Figure 4C–4D). RAF1 amplification potentially resulted from repressed RAD51 expression by EZH2 as co-expression of RAD51 blocked activation of p-ERK-β-catenin signaling (Figure 4C–4D). Together, these results imply that induced RAF1 amplification and downstream p-ERK-β-catenin activation by EZH2-RAD51 regulation in BTICs could contribute to promoting survival, proliferation and self renewal of the BTICs.
Increased EZH2 expression physiologically enhances RAF1-ERK-β-catenin signaling in BTICs
To further reveal the physiological relevance between EZH2 and RAF1-ERK-β-catenin activation in primary BTICs, we examined the protein levels RAF1, p-ERK and unphosphorylated β-catenin under EZH2 induction by hypoxia in CD44+CD24−/low cells using intracellular staining analysis. Consistent with the results shown in Figure 4D, increased EZH2 expression by hypoxia enhanced expression of RAF1, p-ERK and unphosphorylated β-catenin, whereas knocking-down EZH2 by shRNA significantly reversed the hypoxia-activated RAF1-ERK-β-catenin signaling in CD44+CD24−/low cells (Figure S2A). We found that EZH2 expression positively correlates with endogenous RAF1 and unphosphorylated β-catenin expression in all three BTIC populations from three individual breast tumor samples (Figure S2B). Together, these data support the results obtained in the ectopic expression system (Figure 4D) that increased EZH2 expression in BTICs resulted in activation of RAF1-ERK-β-catenin signaling.
EZH2-induced activation of RAF1-p-ERK-β-catenin signaling is positively correlated with breast cancer progression
To elucidate the clinical relevance of EZH2 and RAF1-ERK-β-catenin activation in breast cancer, we first examined association between endogenous EZH2 and RAF1 levels by re-analyzing cDNA Microarray databases from an established human breast cancer study (Chin et al., 2006). We found that endogenous EZH2 was highly expressed in aggressive high stage (stage 3–4) tumors, where RAF1 expression was significantly correlated with EZH2 expression (Figure 4E). We next asked whether overexpression of RAF1 in those tumors expressing high level of endogenous EZH2 is due to RAF1 amplification at the genomic level. In a cohort of human breast tumor sections, over 60% of high grade (II, III) breast tumors showed high EZH2 expression (Table 1). These tumors with high level of endogenous EZH2 frequently exhibited RAF1 gene amplification (Figure 4F, right). On the contrary, RAF1 amplification did not occur in any of the low grade tumors in which 90% of them showed none or low EZH2 expression (Table 1). Furthermore, EZH2 expression was negatively correlated with RAD51 but positively correlated with RAF1, p-ERK and β-catenin expression in these high grade, poorly-differentiated breast carcinoma by immunohistochemistry analysis (Figure 4F, Figure 4G and Table 2). Together, these data support the notion that EZH2, through downregulating RAD51, induces RAF1 amplification and downstream p-ERK-β-catenin activation, which may contribute significantly to promote BTICs and aggravate breast cancer malignancy.
EZH2-amplified RAF1 signaling enhances survival and proliferation of BTICs
To dissect the dynamics of EZH2-amplified RAF1 signaling in regulating survival of the BTIC population, we monitored RAF1 gene copy number following the time course of mammospheric cell division from single cell suspension enriched for BTICs (Figure S3A). During the first 48 hours after seeding, RAD51 was repressed by EZH2 expression (Figure S3B), and we observed approximately 1.3–1.5 fold more cell death of EZH2-infected mammospheric cells compared to vector-infected mammospheric cells with etoposide treatment (Figure S3C), which is consistent with a previous report showing that repression of RAD51 leads to enhanced cell death (Zeidler et al., 2005). However, we found that the EZH2-infected cells start to show decreased cell death compared to the vector control at 72 hours after seeding (Figure S3C) when RAF1 amplification also evidently occurs (Figure S3D). Additionally, EZH2-infected CD44+CD24−/low cells showed resistance to etoposide induced-cell death (Figure S3E) whereas ectopic expression of EZH2 in non-CD44+CD24−/low led to increased cell death upon etoposide treatment (Figure S3E), indicating distinct differences between stem/progenitor cell enriched-populations and non-stem/progenitor cells. Together, these data suggest that enhanced EZH2 expression may amplify RAF1 signaling in BTICs and play a role in promoting survival of the BTICs.
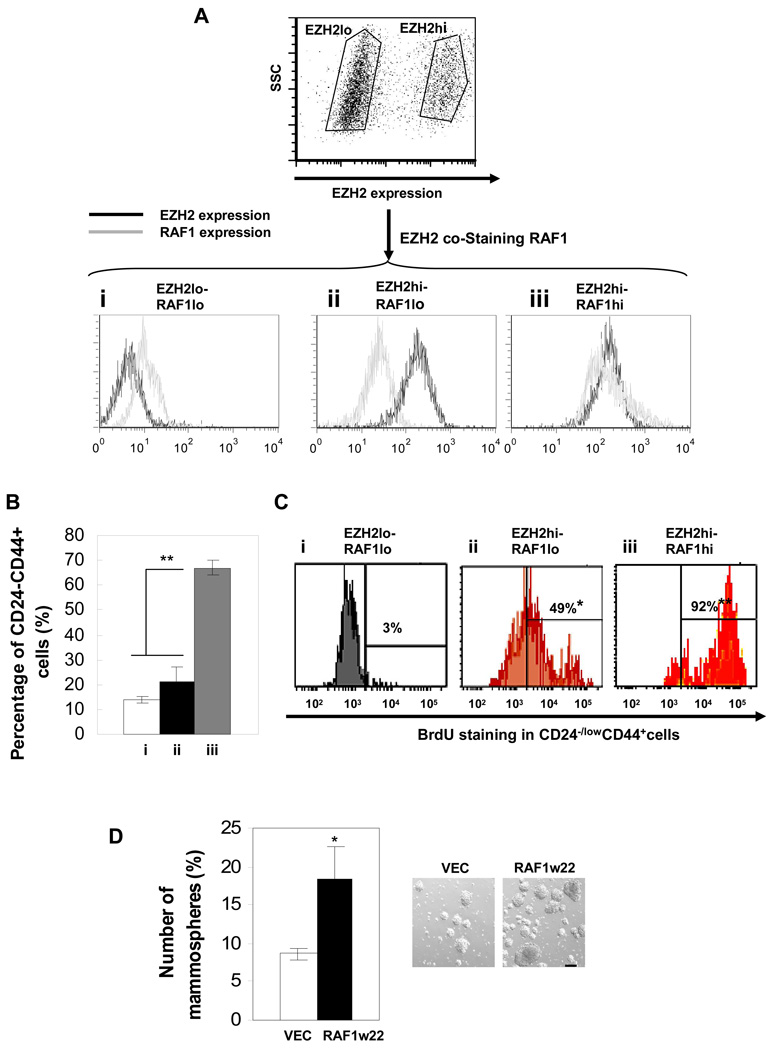
To further assess the biological function of EZH2-amplified RAF1 signaling in regulating BTICs, we isolated three distinct cell populations expressing low EZH2-low RAF1, high EZH2-low RAF1, and high EZH2-high RAF1 (low EZH2-high RAF1 population is absent) from hypoxia treated-primary tumor cells using intracellular staining (Figure 5A). We next determined the proportion of the CD44+CD24−/low cells in each of the three populations and proliferation status of these CD44+CD24−/low cells by BrdU-FITC staining. Among the three populations (Figure 5A), cells expressing high EZH2-high RAF1 had the most abundant CD44+CD24−/low cell population with the highest proliferation rate (BrdU+) (Figure 5B and 5C). Consistent with this data, expression of constitutively activated RAF1 (w22) in human primary tumor cells substantially increased mammosphere number and size (Figure 5D). Together, these data support the notion that amplified RAF1 signaling may not only play a role in promoting BTIC survival but is also essential for enhancing proliferation of BTICs.
Figure 5. EZH2-RAF1 signaling promotes proliferation of BTICs.
(A) Human primary tumor cells were hypoxia treated for 48hrs, cultured in suspension for 3 days, and then co-stained with EZH2 and RAF1 antibodies and secondary antibodies conjugated with APC and PerCp-Cy5.5 respectively to determine the indicated protein expression profiles (i, ii, iii) using flow cytometry (expression pattern: hi: high, lo: low). (B) The percentage of CD44+CD24−/low cells and (C) the percentage of BrdU-FITC positive-CD44+CD24−/low cells for each expression pattern were determined using flow cytometry. (D) Human primary tumor cells expressing constitutively active pBabe-cRAF-w22 (RAF1w22) were cultured as mammospheres, and the number of spheres was counted per 1000 cells (scale bar: 100µm). Error bars denote ±SD (*P<0.05, **P<0.01). See also Figure S3.
A potential model of DSB induced-RAF1 amplification and expansion of RAF1-amplified BTICs
It was shown that repression of RAD51 leads to decreased DNA damage repair and increased double strand breaks (DSB) (van Gent et al., 2001). DSB generates gene amplification through a well-established model- Break-Fusion-Bridge (BFB) cycles (Ford and Fried, 1986). The broken end of the bridge can fuse to sister chromatid during DNA synthesis and starts a fusion-bridge cycle (Figure S3F). Each cycle will generate at least an additional copy of the amplified gene. Given the critical role of RAD51 in DSB repair, we asked whether RAD51 repression could lead to RAF1 amplification through DSB induced-BFB cycles. By using FISH probes targeting RAF1 gene (red) and a co-amplified marker centromeric to RAF1 (green), we found that EZH2-expressed BTICs show inverted duplication of amplified RAF1 gene region, a distinct product from BFB process, suggesting that RAD51 repression leads to RAF1 amplification through DSB induced-BFB cycles (Figure S3G).
To further understand whether rapid amplification of the RAF gene occurs in a small number of cells as a result of having a selective advantage to outgrow the population, or whether there is amplification broadly in the population that somehow specifically targets the RAF gene by an unknown mechanism, we monitored dynamic accumulation of RAF1 amplification in BTICs by FISH (Figure S3H). The number of RAF1-amplified cells from BTIC-enriched primary tumor cells was graphed against the generation number for four generations. We found that RAF1 amplification firstly occurs in a small population of BTICs (3–5% in the first generation) and then expands to ~60% in the fourth generation, which suggests an outgrowth advantage of the RAF1-amplified cell population from EZH2-expressed BTICs (Figure S3I). Since it was shown that HR predominantly repairs DSBs in the S phase and that DSBs caused by HR deficiency are usually replication associated (Branzei et al., 2008), we think the original BTIC population that undergoes replication is likely to be the very first population prone to DSBs and RAF1 amplification. To test this hypothesis, we co-stained Vector-or EZH2-infected BTICs in the first generation with antibodies against BrdU and γ-H2AX, a marker for DSBs. As compared to the Vector-expressed BTICs which showed no DSB formation (Figure S3J, upper panel), spontaneous DSBs caused by EZH2 expression predominantly accumulated in the BrdU-positive cells (Figure S3J, lower panel). Furthermore, there was around 5–10% replicating BTICs in the zero-first generation (Figure S3K), half of which would have DSBs and potentially generate RAF1 amplification through BFB cycles in the first generation. This result is consistent with the percentage of RAF1 amplified population (3–5%) shown in Figure S3I. These 3–5% BTICs carrying EZH2-RAF1 amplification had enhanced proliferation and self-renewal, which enable them to gradually outgrow the other populations (Figure 2, Figure 5 and Figure S3I). Together, our results suggest that RAF1 amplification could be a direct result of EZH2-mediated RAD51 repression through accumulation of DSBs followed by BFB cycles, which may preferentially occur in a replicating BTIC population.
AZD6244 eliminates BTICs and mammary tumor formation by inhibiting RAF1-MEK-ERK-β-catenin activation
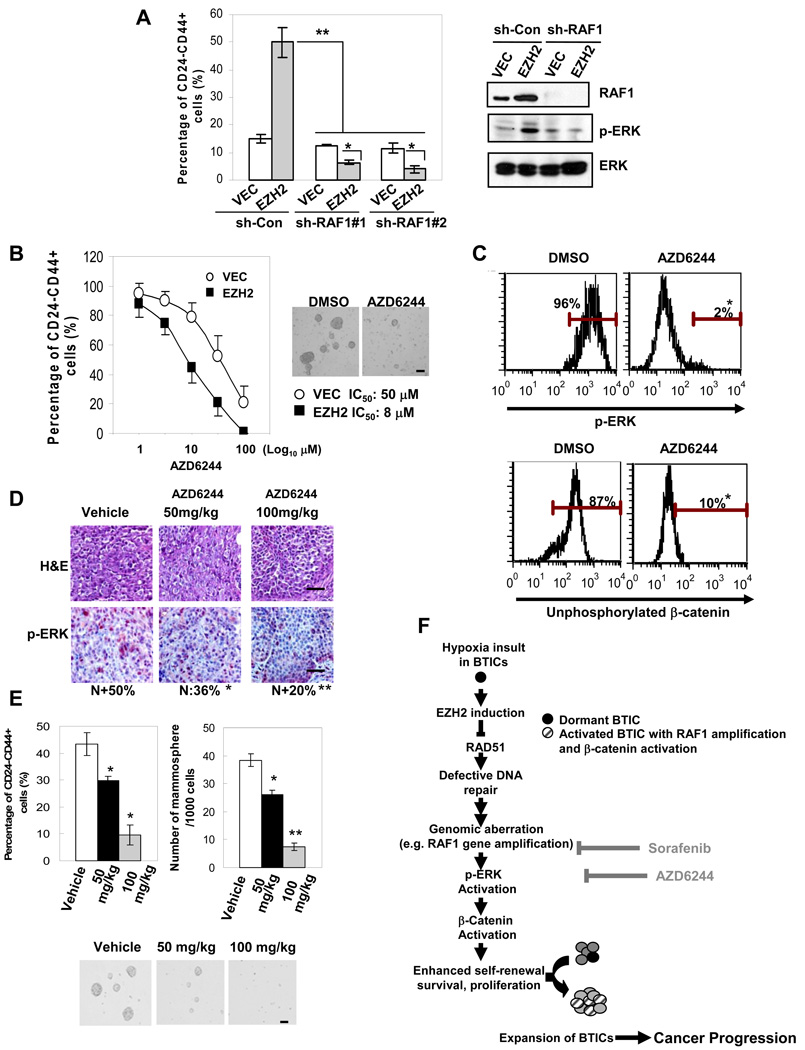
To explore if therapeutic agents that block the RAF-β-catenin activation are able to eliminate EZH2-induced BTIC expansion, we tested sorafenib (Nexavar), a clinically used RAF inhibitor and an FDA-approved anti-cancer drug compared to several clinical cancer therapeutics (imatinib, laptinib, Taxol) and a pharmacological agent that kills cancer cells by activating p53 (Reactivation of p53 and Induction of Tumor Apoptosis, RITA). Sorafenib was the most effective in eliminating CD44+CD24−/low population from BT549 cancer cells (Figure S4A). Sorafenib also significantly decreased CD44+CD24−/low cells from various basal breast cancer cell lines (Figure S4B). Additionally, sorafenib preferentially inhibited EZH2 promoted-BTICs which were under treatment in vitro (Figure S4C–4D) or were isolated from xenograt tumors of the treated mice in vivo (Figure S4E–4F). Sorafenib also abolished EZH2 promoted-mammosphere formation from primary tumor cells as well as from xenograft tumor cells (Figure S4D–4F). Furthermore, knocking-down RAF1 with shRNAs recapitulated the effect of sorafenib in decreasing CD44+CD24−/low BTIC population from human primary tumor cells and also suppressed BTIC xenograft tumor formation (Figure 6A and Table S3). These data suggest that the RAF1 activation pathway is ubiquitously shared and functionally important in EZH2-promoted BTICs. To block ERK activation, we used a specific MEK/ERK inhibitor AZD6244 that has been tested in multiple clinical trials. We found that AZD6244 preferentially eliminates EZH2-expressed CD44+CD24−/low cells and mammospheres (Figure 6B). Furthermore, AZD6244 significantly decreased p-ERK (Figure 6C) and unphosphorylated β-catenin (Figure 6C), and also abolished xenograft tumor formation generated from the primary BTICs that had been pretreated with AZD6244 (Table S3). To determine if AZD6244 also affects BTICs in vivo, we applied AZD6244 to the mice bearing xenograft tumors that were generated from the primary BTICs highly expressing EZH2. Consistent with the in vitro results, AZD6244 inhibited p-ERK (Figure 6D), suppressed CD44+CD24−/low cells (Figure 6E, left), and abolished formation of mammospheres (Figure 6E, right and lower panel) which were recovered from the xenograft tumors in a dose-dependent manner compared to the vehicle treated group. All these data suggest that activation of RAF1-ERK-β-catenin signaling likely plays a role in promoting BTIC expansion and that inhibiting RAF1-MEK-ERK-β-catenin activation by AZD6244 or sorafenib eliminates EZH2-promoted BTICs.
Figure 6. AZD6244 significantly eliminates BTICs and mammary xenograft tumor formation by inhibiting RAF1-MEK-ERK-β-catenin activation.
(A) Protein expression change in BTIC enriched-human primary tumor cells expressing RAF1-shRNAs was examined using Immunoblotting, and the percentage of CD44+CD24−/low cells was determined using flow cytometry. (B) Bar graph showing the percentage of CD44+CD24−/low population from human primary tumor cells treated with different doses of AZD6244. Surviving mammospheres were counted after being treated with different doses of AZD6244 for 48 hrs, and shown as representative microscopy images (10µM AZD6244) (scale bar: 100µm). (C) Bars showing percentage of p-ERK and unphosphorylated β-catenin expression in CD44+CD24−/low cells expressing EZH2 with or without 10µM AZD6244 using intracellular staining. (D) NOD/SCID mice (n=5/group) inoculated with CD44+CD24−/low cells isolated from human primary tumor cells expressing EZH2. Mice bearing tumors for 2 weeks (tumor size~50mm3) were treated with the indicated doses of AZD6244 daily for 1 week. Tumor sections were subjected to H&E and p-ERK staining (N: nuclear staining, n=20/tumor) (scale bar: 20µm). (E) Cells were isolated from the xenograft tumors and cultured as mammospheres. The percentage of CD44+CD24−/low cells was determined using flow cytometry (left). The number of mammospheres counted per 1000 cells (right) and shown as representative microscopy images below (scale bar: 100µm). Error bars denote ±SD (*P<0.05, **P<0.01). (F) Illustration showing RAD51 is down-regulated by EZH2 in BTIC-enriched population, leading to impaired DNA damage repair and accumulation of oncogenic hits that promote BTICs (e.g. RAF1 amplification). Targeting the RAF1-ERK-β-catenin activation pathway with therapeutic reagents like AZD6244 and sorafenib can be effective for eliminating BTICs. See also Figure S4 and Table S3.
Discussion
Our study reveals a mechanism in which EZH2 promotes BTIC expansion and cancer progression. We found that RAD51 downregulation by EZH2 leads to impaired DNA damage repair and accumulation of oncogenic hits in favor of promoting BTICs, such as RAF1-β-catenin activation in BTICs (Figure 6F). These findings suggest that targeting the EZH2/Polycomb complex and its downstream activation pathways, such as RAF1-ERK signaling by AZD6244, may be effective for eliminating BTICs and intervening breast cancer recurrence.
The long-lived stem/progenitor cells can be a reservoir for accumulation of the genetic or epigenetic events required for oncogenic transformation (Rossi et al., 2008). Unrepaired DNA damage likely allows sufficient oncogenic hits to accrue in stem/progenitor cells and their progeny. Although it was reported that DNA damage leads to stem/progenitor cell senescence by activating tumor suppressors such as p16INK4a (Rossi et al., 2008), DNA damage induced by EZH2 overexpression likely bypasses this defense mechanism through polycomb-mediated silencing of p16INK4a and p19Arf expression (Bracken et al., 2007; Tzatsos and Bardeesy, 2008), and thereby prevents EZH2 expanded-BTICs from exhaustion. Moreover, cancer stem/progenitor cells show enhanced regulation of ROS scavenging (Diehn et al., 2009) and anti-apoptosis (Konopleva et al., 2002) compared to non-stem cell populations. Therefore, RAF1 amplification induced by RAD51 downregulation may be a "necessary but not sufficient" effect of RAD51 on BTICs, and these compensatory mechanisms might help sustain stem/progenitor cell survival under DNA damage and genomic instability induced by EZH2 overexpression, which could further lead to deregulated signaling changes enabling BTICs to outgrow and expand.
HIF mRNA was shown to be upregulated in the stem cell population from glioma tumors compared to the non-stem cell population (Li et al., 2009), which may provide an explanation for our observation that EZH2 is highly expressed and induced by hypoxia in the BTIC population compared to non-BTICs. Moreover, it was indicated that RAD51 repression by hypoxia is mediated by repressive E2F4/p130 complexes that bind to the proximal promoter of the gene (Bindra et al, 2006). Consistent with this, in our study, we show that EZH2 binds to RAD51 proximal promoter which contains a putative Polycomb Response Element (Figure 1F and Figure S1D). Moreover, we found that E2F4 binds to EZH2 by immunoprecipitation analysis (data not shown), which suggest that both EZH2 and E2F4 may mediate RAD51 repression as a big repressive complex.
Though DNA repair response or expression of certain DNA damage response/ repair proteins have been reported to be elevated in cancer stem cells from a couple different models (Bao et al., 2006; Zhang et al., 2008), few of these molecules have been functionally characterized. In addition, different from RAD51 mediated-”error-free” DNA repair mechanism, the increased DNA damage repair capacity in the above-mentioned studies might be mediated through enhanced “error-prone” DNA damage repair mechanisms, which could in turn to result in chromosomal aberrations and genomic instability (Aguilera and Gomez-Gonzalez, 2008; Rassool et al., 2007). Furthermore, in support of our study, loss of a DSB repair protein BRCA1 has recently been shown to expand mammary luminal progenitor cells (Liu et al., 2008). RAD51 is the crucial partner of BRCA1 for mediating the error-free DNA damage repair by homologous recombination (van Gent et al., 2001). Further studies needed to be carried out in order to elucidate the relationship between various DNA repair mechanisms/molecules and the regulation of BTICs.
The functional association between these amplicons in BTICs is currently under investigation. Although genomic alterations have been observed in a wide range of human diseases, genomic alterations that occur in stem/progenitor cells or tumor initiating cells are poorly characterized. Among the genomic loci that are highly amplified by increased expression of EZH2 (Table S3), 12p13.31 gain has been shown to correlate with basal breast cancer (Chin et al., 2006), and 17q23.1 gain is especially frequent in BRCA1 associated breast cancer (Sinclair et al., 2002). Particularly, the RAF1 gene on 3p25.2 was found recurrently amplified or rearranged in multiple human cancers, where genomic aberrant RAF1 gene often associates with cancer aggressiveness and relapse (Hajj et al., 1990). Activation of RAF-MAPK pathway is also critical to inducing EMT and metastasis, traits that are recently associated with stem cell properties (Thiery et al., 2009). Furthermore, overexpression of wild-type RAF1 is sufficient to induce tumorgenesis in vivo (Kerkhoff et al., 2000), suggesting expression level of RAF1 is a critical parameter for tumor initiation and development. Based on our and others’ observations, more than 50% of aggressive breast tumors show EZH2 overexpression (Kleer et al., 2003, Table 1), and more than one-third of these tumors exhibit RAF1 amplification (Figure 4F), which potentially mark the tumors with high content of BTICs and aggressive malignancy. Consistent with the studies showing that high grade tumors are enriched in high content of TICs (Pece et al.), our results suggest that EZH2 amplified-RAF1 signaling may play a role in expanding an aggressive sub-population of BTICs which promotes cancer progression. Therefore, our study reveals an important clinical implication of using RAF1/MEK/ERK inhibitor in single agent or in combination for intervening cancer progression and recurrence.
Experimental Procedures
Cell culture
Isolation and culture of mammospheres from multiple primary human breast cancer samples or xenograft tumors were performed as described in Ponti et al. and Dontu et al. Differentiated cell cultures were kept in DMEM/F12 supplemented with 10% fetal bovine serum on collagen coated dish (BD Biosciences). Primary tumor samples were acquired from breast cancer patients with informed consent and experimental procedures were conducted with approval by the University of Texas MD Anderson Institutional Review Board.
ChIP analysis
Chromatin immunoprecipitations were modified from the EZ-CHIP (Upstate) protocol using antibodies: anti-HIF1α (Abcam), anti-EZH2 (Zymed), anti-SUZ12 (Abcam), anti-Bmi1 (Abcam) and anti-H3K27Me3 (Upstate). For sequential ChIP: the first elute immunoprecipitated by anti-EZH2 was further immunoprecipitated by anti-H3K27Me3 and IgG antibodies. The percentage of the bound DNA was quantified against the original DNA input. Primer sequences used for ChIP-PCR are as follows: RAD51-5’cccccggcataaagtttgaat3’ (forward); 5’gaagcgccgcactctcctta3’ (reverse) (PCR product containing polycomb response element, see Figure S1D). GAPDH promoter was used as a negative control. For details, please see Supplemental Experimental Procedures.
FACS analysis
Cells were dissociated with trypsin, washed, and resuspended in PBS as single-cell suspension. Cells were then stained with indicated dye/antibodies: Hoechst 33342 (sigma), Anti-CD24-PE (BD Pharmingen), Anti-CD44-FITC and CD44-APC (BD Pharmingen). To analyze activated β-catenin, cells were stained with surface markers (CD24 and CD44), fixed, permeabilized, and then stained with anti-unphosphorylated β-catenin (8E4) (Axxora), anti-EZH2 (Cell Signaling), anti-p-ERK (Cell Signaling), RAF1, γ-H2AX, BrdU-FITC, BrdU-PE (BD Transduction Laboratories). These cells were washed twice and then stained with secondary antibodies conjugated with APC or PerCP-Cy5.5 (Jackson ImmunoResearch). The immunoflourescence was evaluated by BD FACS Canto II (BD Immunocytometry Systems) and analyzed by FCS3 Express Research Lite Edition (Denovo Software). Cell sorting was performed by BD FACS Aria.
Mammary xenograft tumor
Mammary fat pads of female NOD/SCID mice were inoculated with indicated number of isolated CD44+CD24−/low cells or non-CD44+CD24−/low cells for a total volume of 100 µl per injection site. After 12 weeks, all of the tumors were calculated and animals were sacrificed. For the in vivo treatment, AZD6244 or sorafanib prepared as described in the previous studies (Huynh et al.) was given orally to the mice that had been inoculate with 105 CD44+CD24−/low cells with the indicated doses when the tumors were grown for 2 weeks (tumor size 50mm3). All surgical procedures and animal manipulations were performed under regulations of Division of Laboratory Animal Medicine at The University of Texas MD Anderson Cancer Center. Animal protocols (#06–87–06138) were reviewed and approved by the Institutional Animal Care and Use Committee at The University of Texas MD Anderson Cancer Center.
IHC staining and Fluorescence in situ hybridization (FISH) analysis
Immunohistochemical staining was performed as previously described (Ding et al., 2005). FISH was performed on paraffin embedded breast tumor slides or freshly isolated BTICs following standard protocols. A RAF1 plasmid labeled with green fluorescence by nick-translation was used as probe. RAF1 signal (green) to CEP3 (red) signal ratio>2 was considered as amplification. For assaying inverted duplication, BAC clones covering RAF1 gene (RP11-275J11, red) and Co-amplified marker centromeric to RAF1 (RP11-767C1, green) were used as probes.
Antibody array
The Panorama Ab Microarray-Cell Signaling was purchased from Sigma-Aldrich. Microarray images were analyzed with the GenePixTM Pro 4.0 image analysis software. Fluorescence intensity measurements were normalized with local background, and cytoskeleton antibodies (actins, tubulins) were used for normalization of total protein quantity between samples.
SNP array
SNP array analysis was performed using Affymetrix 6.0 arrays and the data were analyzed and plotted by Partek Genomic Suite (Partek Incorporated). All vector control samples were examined against HapMap normal data sets (Affymetrix) and no RAF1 amplification was observed in any of them.
Accession number
SNP array data are deposited in GEO archive (GSE24144).
Statistical analysis
All data were presented as means± the standard deviation of the mean (SD). Statistical calculations were performed with Microsoft Excel analysis tools. Differences between individual groups were analyzed by paired t test. P values of <0.05 considered statistically significant.
Significance
Tumor initiating cells are suggested to be responsible for initiation and aggressive progression of cancer. We show that enhanced expression of Polycomb protein EZH2 in BTICs (breast tumor initiating cells) results in downregulation of DNA damage repair protein and accumulation of genomic abnormalities that mediate deregulated signaling (RAF1-ERK-β-catenin) promoting BTIC expansion and cancer progression. Our findings also indicate that the cancer-predisposed microenvironment, such as hypoxia, may promote BTICs through upregulating EZH2 expression. Our data further provide evidences linking a specific genomic aberration mediated by EZH2-impaired DNA damage response to expansion of BTICs. Together, this study reveals a previously unidentified therapeutic effect of RAF1-ERK signaling inhibitors to prevent breast cancer progression by eliminating BTICs and elucidates important clinical implications.
Supplementary Material
Acknowledgements
We thank Dr. Peter Glazer for RAD51-shRNA and RAD51-Luciferase constructs, Drs. Phoebus Shiaw-Yih Lin and Peng Guang for active discussion and suggestions, and Drs. Stephanie A. Miller, Jennifer L. Hsu, and Jeng C. Cheng for editing the manuscript. This work was supported by NIH RO1 CA109311, Breast SPORE CA116199, CCSG CA16672, DOD COE W81WXH-06-2-0033, NSC-96-3111-B (NSC, Taiwan), National Breast Cancer Foundation, Inc., Kadoorie Charitable Foundation, Breast Cancer Research Foundation, M.D. Anderson-China Medical University and Hospital Sister Institution Fund and in memoriam, Mrs. Serena Lin-Guo for her courageous fight for breast cancer.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat Rev Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Clarke MF. Self-renewal and solid tumor stem cells. Oncogene. 2004;23:7274–7282. doi: 10.1038/sj.onc.1207947. [DOI] [PubMed] [Google Scholar]
- Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bao S, Wu Q, McLendon RE, Hao Y, Shi Q, Hjelmeland AB, Dewhirst MW, Bigner DD, Rich JN. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature. 2006;444:756–760. doi: 10.1038/nature05236. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Crosby ME, Glazer PM. Regulation of DNA repair in hypoxic cancer cells. Cancer Metastasis Rev. 2007;26:249–260. doi: 10.1007/s10555-007-9061-3. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Glazer PM. Repression of RAD51 gene expression by E2F4/p130 complexes in hypoxia. Oncogene. 2007;26:2048–2057. doi: 10.1038/sj.onc.1210001. [DOI] [PubMed] [Google Scholar]
- Bindra RS, Schaffer PJ, Meng A, Woo J, Maseide K, Roth ME, Lizardi P, Hedley DW, Bristow RG, Glazer PM. Down-regulation of Rad51 and decreased homologous recombination in hypoxic cancer cells. Mol Cell Biol. 2004;24:8504–8518. doi: 10.1128/MCB.24.19.8504-8518.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bracken AP, Kleine-Kohlbrecher D, Dietrich N, Pasini D, Gargiulo G, Beekman C, Theilgaard-Monch K, Minucci S, Porse BT, Marine JC, et al. The Polycomb group proteins bind throughout the INK4A-ARF locus and are disassociated in senescent cells. Genes Dev. 2007;21:525–530. doi: 10.1101/gad.415507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bristow RG, Hill RP. Hypoxia and metabolism. Hypoxia, DNA repair and genetic instability. Nat Rev Cancer. 2008;8:180–192. doi: 10.1038/nrc2344. [DOI] [PubMed] [Google Scholar]
- Cao R, Zhang Y. The functions of E(Z)/EZH2-mediated methylation of lysine 27 in histone H3. Curr Opin Genet Dev. 2004;14:155–164. doi: 10.1016/j.gde.2004.02.001. [DOI] [PubMed] [Google Scholar]
- Chen MS, Woodward WA, Behbod F, Peddibhotla S, Alfaro MP, Buchholz TA, Rosen JM. Wnt/beta-catenin mediates radiation resistance of Sca1+ progenitors in an immortalized mammary gland cell line. J Cell Sci. 2007;120:468–477. doi: 10.1242/jcs.03348. [DOI] [PubMed] [Google Scholar]
- Chin K, DeVries S, Fridlyand J, Spellman PT, Roydasgupta R, Kuo WL, Lapuk A, Neve RM, Qian Z, Ryder T, et al. Genomic and transcriptional aberrations linked to breast cancer pathophysiologies. Cancer Cell. 2006;10:529–541. doi: 10.1016/j.ccr.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Diehn M, Cho RW, Lobo NA, Kalisky T, Dorie MJ, Kulp AN, Qian D, Lam JS, Ailles LE, Wong M, et al. Association of reactive oxygen species levels and radioresistance in cancer stem cells. Nature. 2009;458:780–783. doi: 10.1038/nature07733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Difilippantonio MJ, Petersen S, Chen HT, Johnson R, Jasin M, Kanaar R, Ried T, Nussenzweig A. Evidence for replicative repair of DNA double-strand breaks leading to oncogenic translocation and gene amplification. J Exp Med. 2002;196:469–480. doi: 10.1084/jem.20020851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Xia W, Liu JC, Yang JY, Lee DF, Xia J, Bartholomeusz G, Li Y, Pan Y, Li Z, et al. Erk associates with and primes GSK-3beta for its inactivation resulting in upregulation of beta-catenin. Mol Cell. 2005;19:159–170. doi: 10.1016/j.molcel.2005.06.009. [DOI] [PubMed] [Google Scholar]
- Dodson H, Bourke E, Jeffers LJ, Vagnarelli P, Sonoda E, Takeda S, Earnshaw WC, Merdes A, Morrison C. Centrosome amplification induced by DNA damage occurs during a prolonged G2 phase and involves ATM. Embo J. 2004;23:3864–3873. doi: 10.1038/sj.emboj.7600393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, Wicha MS. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003;17:1253–1270. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ezhkova E, Pasolli HA, Parker JS, Stokes N, Su IH, Hannon G, Tarakhovsky A, Fuchs E. Ezh2 orchestrates gene expression for the stepwise differentiation of tissue-specific stem cells. Cell. 2009;136:1122–1135. doi: 10.1016/j.cell.2008.12.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo W, Lasky JL, Chang CJ, Mosessian S, Lewis X, Xiao Y, Yeh JE, Chen JY, Iruela-Arispe ML, Varella-Garcia M, Wu H. Multi-genetic events collaboratively contribute to Pten-null leukaemia stem-cell formation. Nature. 2008;453:529–533. doi: 10.1038/nature06933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajj C, Akoum R, Bradley E, Paquin F, Ayoub J. DNA alterations at proto-oncogene loci and their clinical significance in operable non-small cell lung cancer. Cancer. 1990;66:733–739. doi: 10.1002/1097-0142(19900815)66:4<733::aid-cncr2820660422>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- Hu M, Polyak K. Molecular characterisation of the tumour microenvironment in breast cancer. Eur J Cancer. 2008;44:2760–2765. doi: 10.1016/j.ejca.2008.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh H, Ngo VC, Koong HN, Poon D, Choo SP, Toh HC, Thng CH, Chow P, Ong HS, Chung A, et al. AZD6244 enhances the anti-tumor activity of sorafenib in ectopic and orthotopic models of human hepatocellular carcinoma (HCC) J Hepatol. 52:79–87. doi: 10.1016/j.jhep.2009.10.008. [DOI] [PubMed] [Google Scholar]
- Kerkhoff E, Fedorov LM, Siefken R, Walter AO, Papadopoulos T, Rapp UR. Lung-targeted expression of the c-Raf-1 kinase in transgenic mice exposes a novel oncogenic character of the wild-type protein. Cell Growth Differ. 2000;11:185–190. [PubMed] [Google Scholar]
- Kleer CG, Cao Q, Varambally S, Shen R, Ota I, Tomlins SA, Ghosh D, Sewalt RG, Otte AP, Hayes DF, et al. EZH2 is a marker of aggressive breast cancer and promotes neoplastic transformation of breast epithelial cells. Proc Natl Acad Sci U S A. 2003;100:11606–11611. doi: 10.1073/pnas.1933744100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konopleva M, Zhao S, Hu W, Jiang S, Snell V, Weidner D, Jackson CE, Zhang X, Champlin R, Estey E, et al. The anti-apoptotic genes Bcl-X(L) and Bcl-2 are over-expressed and contribute to chemoresistance of non-proliferating leukaemic CD34+ cells. Br J Haematol. 2002;118:521–534. doi: 10.1046/j.1365-2141.2002.03637.x. [DOI] [PubMed] [Google Scholar]
- Lessard J, Sauvageau G. Polycomb group genes as epigenetic regulators of normal and leukemic hemopoiesis. Exp Hematol. 2003;31:567–585. doi: 10.1016/s0301-472x(03)00081-x. [DOI] [PubMed] [Google Scholar]
- Li Y, Welm B, Podsypanina K, Huang S, Chamorro M, Zhang X, Rowlands T, Egeblad M, Cowin P, Werb Z, et al. Evidence that transgenes encoding components of the Wnt signaling pathway preferentially induce mammary cancers from progenitor cells. Proc Natl Acad Sci U S A. 2003;100:15853–15858. doi: 10.1073/pnas.2136825100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Bao S, Wu Q, Wang H, Eyler C, Sathornsumetee S, Shi Q, Cao Y, Lathia J, McLendon RE, et al. Hypoxia-inducible factors regulate tumorigenic capacity of glioma stem cells. Cancer Cell. 2009;15:501–513. doi: 10.1016/j.ccr.2009.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Ginestier C, Charafe-Jauffret E, Foco H, Kleer CG, Merajver SD, Dontu G, Wicha MS. BRCA1 regulates human mammary stem/progenitor cell fate. Proc Natl Acad Sci U S A. 2008;105:1680–1685. doi: 10.1073/pnas.0711613105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrawala L, Calhoun T, Schneider-Broussard R, Zhou J, Claypool K, Tang DG. Side population is enriched in tumorigenic, stem-like cancer cells, whereas ABCG2+ and ABCG2- cancer cells are similarly tumorigenic. Cancer Res. 2005;65:6207–6219. doi: 10.1158/0008-5472.CAN-05-0592. [DOI] [PubMed] [Google Scholar]
- Pece S, Tosoni D, Confalonieri S, Mazzarol G, Vecchi M, Ronzoni S, Bernard L, Viale G, Pelicci PG, Di Fiore PP. Biological and molecular heterogeneity of breast cancers correlates with their cancer stem cell content. Cell. 140:62–73. doi: 10.1016/j.cell.2009.12.007. [DOI] [PubMed] [Google Scholar]
- Ponti D, Costa A, Zaffaroni N, Pratesi G, Petrangolini G, Coradini D, Pilotti S, Pierotti MA, Daidone MG. Isolation and in vitro propagation of tumorigenic breast cancer cells with stem/progenitor cell properties. Cancer Res. 2005;65:5506–5511. doi: 10.1158/0008-5472.CAN-05-0626. [DOI] [PubMed] [Google Scholar]
- Rassool FV, Gaymes TJ, Omidvar N, Brady N, Beurlet S, Pla M, Reboul M, Lea N, Chomienne C, Thomas NS, et al. Reactive oxygen species, DNA damage, and error-prone repair: a model for genomic instability with progression in myeloid leukemia? Cancer Res. 2007;67:8762–8771. doi: 10.1158/0008-5472.CAN-06-4807. [DOI] [PubMed] [Google Scholar]
- Reya T, Duncan AW, Ailles L, Domen J, Scherer DC, Willert K, Hintz L, Nusse R, Weissman IL. A role for Wnt signalling in self-renewal of haematopoietic stem cells. Nature. 2003;423:409–414. doi: 10.1038/nature01593. [DOI] [PubMed] [Google Scholar]
- Reya T, Morrison SJ, Clarke MF, Weissman IL. Stem cells, cancer, and cancer stem cells. Nature. 2001;414:105–111. doi: 10.1038/35102167. [DOI] [PubMed] [Google Scholar]
- Rossi DJ, Jamieson CH, Weissman IL. Stems cells and the pathways to aging and cancer. Cell. 2008;132:681–696. doi: 10.1016/j.cell.2008.01.036. [DOI] [PubMed] [Google Scholar]
- Sinclair CS, Adem C, Naderi A, Soderberg CL, Johnson M, Wu K, Wadum L, Couch VL, Sellers TA, Schaid D, et al. TBX2 is preferentially amplified in BRCA1- and BRCA2-related breast tumors. Cancer Res. 2002;62:3587–3591. [PubMed] [Google Scholar]
- Sing A, Pannell D, Karaiskakis A, Sturgeon K, Djabali M, Ellis J, Lipshitz HD, Cordes SP. A vertebrate Polycomb response element governs segmentation of the posterior hindbrain. Cell. 2009;138:885–897. doi: 10.1016/j.cell.2009.08.020. [DOI] [PubMed] [Google Scholar]
- Sparmann A, van Lohuizen M. Polycomb silencers control cell fate, development and cancer. Nat Rev Cancer. 2006;6:846–856. doi: 10.1038/nrc1991. [DOI] [PubMed] [Google Scholar]
- Taipale J, Beachy PA. The Hedgehog and Wnt signalling pathways in cancer. Nature. 2001;411:349–354. doi: 10.1038/35077219. [DOI] [PubMed] [Google Scholar]
- Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- Tzatsos A, Bardeesy N. Ink4a/Arf regulation by let-7b and Hmga2: a genetic pathway governing stem cell aging. Cell Stem Cell. 2008;3:469–470. doi: 10.1016/j.stem.2008.10.008. [DOI] [PubMed] [Google Scholar]
- Varambally S, Dhanasekaran SM, Zhou M, Barrette TR, Kumar-Sinha C, Sanda MG, Ghosh D, Pienta KJ, Sewalt RG, Otte AP, et al. The polycomb group protein EZH2 is involved in progression of prostate cancer. Nature. 2002;419:624–629. doi: 10.1038/nature01075. [DOI] [PubMed] [Google Scholar]
- Visvader JE, Lindeman GJ. Cancer stem cells in solid tumours: accumulating evidence and unresolved questions. Nat Rev Cancer. 2008;8:755–768. doi: 10.1038/nrc2499. [DOI] [PubMed] [Google Scholar]
- Wicha MS. Cancer stem cell heterogeneity in hereditary breast cancer. Breast Cancer Res. 2008;10:105. doi: 10.1186/bcr1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeidler M, Varambally S, Cao Q, Chinnaiyan AM, Ferguson DO, Merajver SD, Kleer CG. The Polycomb group protein EZH2 impairs DNA repair in breast epithelial cells. Neoplasia. 2005;7:1011–1019. doi: 10.1593/neo.05472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang M, Behbod F, Atkinson RL, Landis MD, Kittrell F, Edwards D, Medina D, Tsimelzon A, Hilsenbeck S, Green JE, et al. Identification of tumor-initiating cells in a p53-null mouse model of breast cancer. Cancer Res. 2008;68:4674–4682. doi: 10.1158/0008-5472.CAN-07-6353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.