Abstract
With the overwhelming volume of genomic and molecular information available on many databases nowadays, researchers need from bioinformaticians more than encouragement to refine their searches. We present here GeneRanker, an online system that allows researchers to obtain a ranked list of genes potentially related to a specific disease or biological process by combining gene-disease (or genebiological process) associations with protein-protein interactions extracted from the literature, using computational analysis of the protein network topology to more accurately rank the predicted associations. GeneRanker was evaluated in the context of brain cancer research, and is freely available online at http://www.generanker.org.
Introduction
It is an exciting time for genomic research: the Human Genome Project is complete, a wide array of high-throughput gene expression analysis techniques is now available, and genomic data is pouring into public databases. The challenge now is to translate genomic research to improved human health.
In order to discover potential gene candidates for their role in specific disease-related behavior, researchers often use gene lists from different sources (such as genes annotated for a specific function in the Gene Ontology) as a starting point, proceeding to further computational analysis (say by comparing against clinical datasets for differential expression of the target genes) and empirical validation. This process allows some room for discovery, as the function mapping of the Gene Ontology might not be specific to the disease the researcher is studying. However, these findings do not stray too far from established knowledge, creating the bursts of popularity observed for genes that become widely studied at around the same time by many laboratories.
On the other hand, it is difficult for researchers to incorporate little known molecular associations and new knowledge in their discovery process, given the impressive volume of publications that need to be reviewed. For example, a search for a single gene, TNF alpha, yields 76,220 articles, with almost half of them published in the last 5 years. Refining the search to TNF alpha and inflammation reduces this number to 15676 articles, still too many for a researcher to review. Finding all the protein-protein interactions in which TNF alpha is involved (in order to discover new potential targets) becomes an even more frustrating exercise, as there is no way to express such a query in PubMed, and existing curated interaction databases cover a very small fraction of the existing relevant literature1 (less than 5%(1)).
It has become a self-evident truth that genetic researchers need more from bioinformaticians than encouragement to refine their searches. The volume of information requires advanced analysis and integration techniques that need to yield trustworthy results of biological relevance. We present GeneRanker, an online system that allows researchers to obtain a ranked list of genes potentially related to a disease or biological process which integrates knowledge from the literature with graph-theoretical analysis of relevant protein-protein interactions.
Background
Data sources
GeneRanker serves as an interface to a method for predicting gene-disease associations by combining data extracted from the literature and from curated sources. Its biological and mathematical basis were introduced in (2). GeneRanker is not an information extraction system nor a natural language processing system, as it relies on data stored by another system from our lab, CBioC (3). The CBioC database contains over 1 million protein-protein interactions as well as over 300,000 gene-disease and 250,000 gene-biological process associations extracted from 1.6 million biomedical abstracts using natural language processing2. Details on related NLP work have been previously reported. CBioC also integrates close to 380,000 protein-protein interactions from IntAct(4), MINT(5), BIND(6), and DIP(7). The NLP engine behind CBioC, IntEx, has been rated as around 65% accurate for protein-protein extractions from text (8), and at 77% for gene-disease association extractions. We sought to increase the precision of the extracted data through knowledge integration via the ranking method presented in (2), briefly described next.
Method
The knowledge integration and data analysis method behind the GeneRanker interface can be summarized as follows:
Given a target disease or biological process name, obtain a list of genes known to be involved with the target disease from the CBioC database. This becomes the initial set.
Extract from the CBioC database all interactions that involve genes in the initial set. A network where each protein has edges to others with which it reportedly interacts is built from this set. Edges are weighted with a “confidence level” that reflects the source of the interactions. Interactions extracted from the literature have a confidence of 0.65 (the reported accuracy of the NLP engine behind CBioC) and those from curated sources a 1.0. The proteins in this network form the extended set.
Apply a two-part scoring formula to each the extended set to predict the proteins most likely related to the disease.
The first part of the scoring formula counts the number of interactions of each protein in the extended set with proteins in the initial set, and weights it with the average confidence of the interactions. The second part of the score measures the importance of the protein in keeping its “neighborhood” connected. Given that high degrees of local network interconnectivity identify sets of functionally related proteins (9, 10), we hypothesized that the relative importance of a protein in keeping this connectivity could reflect its biological relevance for particular molecular behaviors, and by extension (as the network is derived from proteins that are potentially relevant to a specific disease), its biological relevance with respect to the disease. The two measures are then combined using their harmonic mean.
To measure the connectivity of a protein p, we use the traditional clustering coefficient measure from graph theory: the ratio of actual edges in the neighborhood of p to the maximum number of edges that can exist in the neighborhood (11), denoted cc(p). If cc(p) is close to 1, the small set formed by p and its neighbors is highly connected, and thus, if p were to be “removed” from the network (aberrantly expressed), there would likely be another “connection” around its neighbors, and the network will remain connected (and the local function preserved). Otherwise, its “absence” will likely affect the function of the cluster, as there are not too many alternative paths. To be able to combine it with the first part of the scoring formula, we use 1-cc(p) as the second part of the score for p (to match the implication of importance derived from a high/low score).
GeneRanker Interface
The current implementation of GeneRanker includes the basic information for the method to work. Options that will allow users to add weights to the scoring process (such as adding to the score of interactions due to phosphorylation, or to those that involve genes in a certain region of the genome), as well as gene/protein annotation features are in development.
Selecting the disease to study
GeneRanker allows users to enter a search term, which can be a disease or biological process –such as glioblastoma or apoptosis-, and then queries the CBioC database for a list of genes and proteins found to be associated with that disease or process.
Working with the initial set of genes
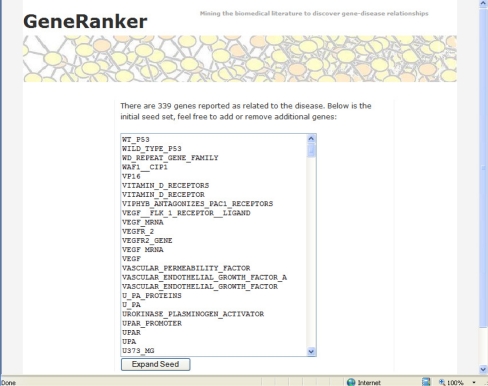
The list of genes or proteins found to be associated with the disease or biological process is displayed in full (Figure 1). Depending on the term, there can be anywhere from 20 to over 300 genes and proteins in the list. The user can add other genes to this initial set or remove any deemed to be redundant or incorrect. Once the initial set is finalized, the protein network will be constructed upon clicking on the “Expand the Network” button.
Figure 1.
Initial set of genes. After the user types a disease or biological process, the GeneRanker system web interface (available at www.generanker.org) displays an initial set of genes obtained from relevant gene-disease associations extracted by CBioC from biomedical literature. Accuracy is estimated at 75% for this initial set.
Ranking the extended set of proteins
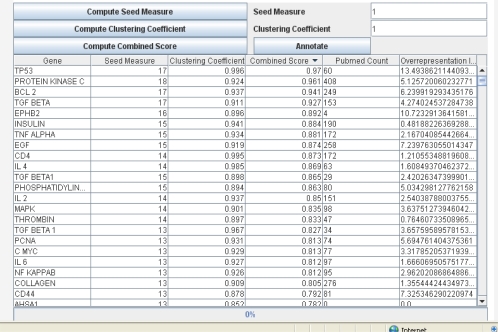
For each gene in the initial set, GeneRanker obtains a set of all the interactions in which it is involved. This is the most time consuming step (taking on average about 10-15 minutes), and results in an extended set of genes that is about 100 times as large as the initial set. All interactions among genes in these larger set (the extended set) form the network that will be analyzed to assign a score to each gene, as described in the previous section. The user can then choose to continue ranking the extended set or store the network (as an XML file). Figure 3 shows the final screen, once the genes have been scored. The user initiates the scoring process by clicking on “Compute Seed Measure”, “Compute Clustering Coefficient” and “Compute Combined Score” in succession. The combined score is the weighted harmonic mean of the two scores, and the user can vary the relative weight of each score by changing the “Seed Measure” and “Clustering Coefficient” values. The process is memory intensive, but usually runs in less than 5 minutes altogether. The user can sort the genes by any of the scores or by gene name.
Figure 3.
GeneRanker, showing the top ranked genes for glioblastoma. The interface allows users to apply the method to any disease or biological process, obtaining a ranked list of potentially related genes. Precision reaches 91 to 94% for the top 100 genes in the list.
Automatic Annotation
A useful feature on this last screen is the “Annotate” function. Users can select a subset of genes which are annotated using information from PubMed. There are two annotation measures: “PubMed Count” and “Overrepresentation Index”. “PubMed Count”, is the number of publications found in PubMed when querying for the protein name and the disease or biological process term together. The “Overrepresentation Index”, calculated as in (12), is a measure of how much more likely it is to find the protein in PubMed together with the diseaserelated term as compared to with other terms. In other words, it is a measure of specificity: it shows the strength of the relationship between a gene and a disease by indicating how much the observed number of co-occurrences in the PubMed documents deviates from the expected number if the cooccurrence were by chance. Thus, an index greater than 1 indicates the cooccurrence is not likely by chance, and the more it exceeds 1, the stronger the association. For example, for “tp53”, “PubMed Count” is 60, while “insulin” has 190. However, the overrepresentation index for tp53 is 13.49, while for insulin, it is 0.48, indicating insulin occurs often with other terms as well, not only glioblastoma. Tempting as it might be to use the overrepresentation index as part of the ranking criteria, it would only help to find known targets, and will exclude potentially valuable new discoveries that can be unveiled through the GeneRanker method. Thus, the measure is included only as aggregated information for the researcher.
Evaluation
We conducted an evaluation of the gene ranking method in the context of a specific disease (glioma), using two different approaches: (i) comparing GeneRanker lists to those obtained from text extraction, and (ii) evaluating GeneRanker results against a clinical glioma dataset.
Comparing GeneRanker to text extraction
Given that biomedical text mining has reached a point were performance improvements of even 1 to 2 percent are very difficult to achieve (and highly significant), a comparison of the precision of GeneRanker to lists obtained from the CBioC database provides a measure of the value of post-processing results from text extraction using the knowledge integration and computational analysis techniques in GeneRanker. The value of such post-processing becomes clear if we compare the 17% performance gain of GeneRanker over text extraction (see Table 1) to the 1.23% that was consider a “highly significant difference” in the Biocreative II gene mention task (13). An overview of current challenges and limitations of biomedical text mining and why traditional extraction techniques are reaching their performance limit for many tasks appears in (14).
Table 1.
Results of comparing GeneRanker to text extraction in finding genes associated to a specific disease. True positives (TP) are genes that are either associated to the disease in OMIM or that were found to co-occur in PubMed abstracts with an overrepresentation index greater than 1. GeneRanker exceeds the performance of text extraction by up to 17%.
| TP | FP | Precision% | |
|---|---|---|---|
| Text extraction list 1 | 153 | 47 | 77% |
| Text extraction list 2 | 159 | 41 | 80% |
| Text extraction list 3 | 150 | 50 | 75% |
| Average(std dev) | 154.0(4.6) | 46.0(4.6) | 77%(2%) |
| GeneRanker (top 50) | 47 | 3 | 94% |
| Effect (gain in precision wrt text extraction) | 17% | ||
| GeneRanker (top 100) | 91 | 9 | 91% |
| Effect (gain in precision wrt text extraction) | 14% | ||
| GeneRanker (top 200) | 175 | 25 | 88% |
| Effect (gain in precision wrt text extraction) | 11% | ||
To measure the performance of a text extraction system such as the one behind CBioC, one rates its precision (percentage of the extracted entities that are considered correct), and its recall (percentage of the available entities extracted). The two measures are often combined using their harmonic mean, or f-measure. In biological domains, and particularly for gene ranking systems, precision is much more relevant than recall(15), as researchers won’t usually mind not getting everything that can possibly be extracted as long as what is extracted is correct. This view was confirmed by the cancer researchers in the team. In view of this, we designed the evaluation methodology to emphasize precision.
In order to compare the precision of the top 200 genes in the GeneRanker list to that of pure text extraction, we extracted all gene-disease associations in the CBioC database where the disease term was either “glioma”, “glioblastoma”, or “astrocytoma” (a total of 1560 entries). We then randomly selected 3 groups of 200 genes each for annotation. Each gene in these 3 lists, plus the top 200 genes from GeneRanker was automatically annotated with two numeric measures obtained from PubMed: one indicated the number of articles returned by searching for the gene name co-occurring with “glioma”, “glioblastoma”, or “astrocytoma”. In addition, we also searched PubMed for the occurrence of the gene alone and the glioma-related terms alone. The number of publications returned was noted for each search. The genes were also similarly searched in OMIM.
Based on whether the gene was in OMIM, plus considering the number of publications where the gene co-occurs with the disease-terms and the overrepresentation index, each gene was marked as either a true positive (TP) or false positive (FP). Results are summarized in Table 1. Genes that had no hits in PubMed were considered a false positive, although they could in fact be related to the disease (these are the potential new targets).
Evaluating against a clinical dataset
The next step of contextual evaluation was to test the method in a biological context for its ability to identify potential gene targets (known or not). The method was run for “glioblastoma”, and the final ranked list was analyzed by TGen’s Brain Tumor Unit researchers Armstrong and McDonough. The top 300 genes reported by GeneRanker to be related to glioma were queried against a whole-genome expression microarray (Affimetrix U133 Plus 2.0) using the Repository of Molecular Brain Neoplasia DaTa (REMBRANDT) Database(16), seeking to discern candidate genes which demonstrate variations in expression related to this type of glioma. Similarly, 10 random gene lists and a list generated from Gene Ontology annotations for cell-cell adhesion, a biological process relevant to glioma. Table 2 summarizes the results.
Table 2.
Evaluation against glioblastoma (GBM) dataset. The percentage of probes with 2-fold differential expression (up or down) in the GBM dataset are noted for (a) a set of 10 random lists of 300 genes, (b) a list of genes obtained from gene ontology (GO) annotations for cell-cell adhesion, and (c) the top 300 genes from GeneRanker. The effect size of the later with respect to the GO list (and therefore, wrt the random list) is highly significant.
| % of 2x diff. expr. probes | |
|---|---|
| Random gene list 1 | 16.4% |
| Random gene list 2 | 13.6% |
| Random gene list 3 | 17.4% |
| Random gene list 4 | 14.5% |
| Random gene list 5 | 14.8% |
| Random gene list 6 | 17.9% |
| Random gene list 7 | 11.6% |
| Random gene list 8 | 18.8% |
| Random gene list 9 | 16.1% |
| Random gene list 10 | 14.8% |
| Average(std dev) | 15.6% (2.2%) |
| GO list for cell-cell adhesion | 19.5% |
| Effect size (wrt random list) | 1.2 |
| GeneRanker top 300 list | 28.2% |
| Effect (% difference wrt GO list) | 8.7% |
| Effect size | 4.0 |
As shown in Table 2, the set of probes obtained from the GeneRanker list are 2x differentially expressed 28.2% of the time, which represents an 8.7% greater yield over the Gene Ontology list. This represents an effect-size statistic of 4.0. Effect size is the preferred method of determining both the statistical and clinical significance of the difference between two groups(17, 18), and, as proposed by Cohen (19), it is estimated by the ratio of the mean difference between the two groups divided by the standard deviation of the control group. In the Cohen (19) scale (adjusted for effect size rather than correlation, as done in (20)), anything with an effect size of over 0.8 is large, between 0.5 and 0.8 is moderate, between 0.2 and 0.5 is small, and anything smaller than 0.2 is insubstantial. Thus, the effect size shown for the GeneRanker list is highly significant.
Conclusion
We have presented GeneRanker, an online tool for predicting associations between proteins and diseases using data from the literature. The precision for GeneRanker was measured to be between 91% and 94% for glioblastoma, surpassing the precision of text extraction systems alone. It also outperforms other gene ranking methods, such as the one by Morrison et al (21), which reports a maximum accuracy of less than 90% for their best combination of inputs; and the one by Seki and Mostafa (22) to associate genes and hereditary diseases, which reports an accuracy of 74% for their best prediction. Our results are thus very encouraging, although evaluation of the method for other diseases is still ongoing.
Overall, GeneRanker was judged by the BTU researchers as a promising tool for finding potential gene targets. In contrast to a list obtained from other sources (such as the Gene Ontology), the GeneRanker top-ranked list includes well-known targets (such as P53, EGFR, VCAM1, AKT1, and CD44) increasing confidence on the tool, as well as potentially novel targets (at least one novel target that could have been missed otherwise has already been identified and is currently under empirical validation).
References
- 1.Donaldson I, Martin J, de Bruijn B, Wolting C, Lay V, Tuekam B, et al. PreBIND and Textomy - mining the biomedical literature for protein-protein interactions using a support vector machine. BMC Bioinformatics. 2003;4(1):11. doi: 10.1186/1471-2105-4-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gonzalez G, Uribe JC, Tari L, Brophy C, Baral C.Mining Gene-Disease relationships from Biomedical Literature: Incorporating Interactions, Connectivity, Confidence, and Context Measures Pacific Symposium in Biocomputing2007;Maui, Hawaii2007 [PubMed] [Google Scholar]
- 3.Baral C, Gonzalez G, Gitter A, Teegarden C, Zeigler A.CBioC: beyond a prototype for collaborative annotation of molecular interactions from the literature Computational Systems Bioionformatics Conference2007;San Diego, CALife Sciences Society; 2007 [DOI] [PubMed] [Google Scholar]
- 4.Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, et al. IntAct: an open source molecular interaction database Nucl Acids Res 2004January12004;32suppl_1D452–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.MINT : a Molecular INteractions Database. [cited; Available from: http://mint.bio.uniroma2.it
- 6.Bader G, Betel D, Hogue C. BIND: the Biomolecular Interaction Network Database. Nucleic Acids Res. 2003;31:248–50. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Database of Interacting Proteins (DIP). [cited; Available from: http://dip.doe-mbi.ucla.edu/
- 8.Ahmed S, Chidambaram D, Davulcu H, Baral C. Intex: A syntactic role driven protein-protein interaction extractor for bio-medical text. Proceedings ISMB/ACL Biolink. 2005:54–61. [Google Scholar]
- 9.Rives AW, Galitski T.Modular organization of cellular networks. PNAS 2003February42003;10031128–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.LaCount DJ, Vignali M, Chettier R, Phansalkar A, Bell R, Hesselberth JR, et al. A protein interaction network of the malaria parasite Plasmodium falciparum. Nature. 2005;438(7064):103–7. doi: 10.1038/nature04104. [DOI] [PubMed] [Google Scholar]
- 11.Brohee S, van Helden J. Evaluation of clustering algorithms for protein-protein interaction networks. BMC Bioinformatics. 2006;7(1):488. doi: 10.1186/1471-2105-7-488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Karopka T, Fluck J, Mevissen H-T, Glass A. The Autoimmune Disease Database: a dynamically compiled literature-derived database. BMC Bioinformatics. 2006;7(1):325. doi: 10.1186/1471-2105-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilbur J.BioCreative 2. Gene Mention Task Overview Second BioCreative Challenge Evaluation Workshop2007; Madrid, Spain; 2007
- 14.Zweigenbaum P, Demner-Fushman D, Yu H, Cohen KB.Frontiers of biomedical text mining: current progress Brief Bioinform 2007September12007;85358–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Owen AB, Stuart J, Mach K, Villeneuve AM, Kim S. A Gene Recommender Algorithm to Identify Coexpressed Genes in C. elegans. Genome Res. 2003 2003 Aug 1;13(8):1828–37. doi: 10.1101/gr.1125403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.The Repository of Molecular Brain Neoplasia DaTa (REMBRANDT) Neuro-Oncology Branch of the National Cancer Institute (NCI) and the National Institute of Neurological Disorders and Stroke (NINDS). [Google Scholar]
- 17.Hojat M, Xu G.A Visitor's Guide to Effect Sizes – Statistical Significance Versus Practical (Clinical) Importance of Research Findings Advances in Health Sciences Education 2004September2004;93241–9. [DOI] [PubMed] [Google Scholar]
- 18.Phillip W, Long MD.When Is A Difference Between Two Groups Significant? Internet Mental Health 2005 [cited 2008 January 31]; Available from: http://www.mentalhealth.com/dis-rs/rs-effect_size.html
- 19.Cohen J. Statistical Power Analysis for the Behavioral Sciences. 2 ed. New Jersey: Lawrence Erlbaum; 1988. [Google Scholar]
- 20.Hopkins WG.A New View of Statistics 2004 [cited; Available from: http://www.sportsci.org/resource/stats/
- 21.Morrison J, Breitling R, Higham D, Gilbert D. GeneRank: Using search engine technology for the analysis of microarray experiments. BMC Bioinformatics. 2005;6(1):233. doi: 10.1186/1471-2105-6-233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kazuhiro Seki JM.Discovering Implicit Associations Between Genes and Hereditary Diseases Pacific Symposium on Biocomputing2007; Maui, Hawaii2007316–27. [PubMed] [Google Scholar]