Abstract
In bioinformatics, clinical data is rarely used. Here, we propose using bedsidedata in basic research, via bioinformatics methodologies. To demonstrate the potential of this so called Reverse Translational Bioinformatics approach, classical bioinformatics tools were applied to blood biomarker information attained from a large scale, open-access cross sectional survey. The results of this analysis include a novel classification of blood biomarkers, critical ages in which basic biological processes may shift in humans, and a possible approach to exploring the gender specificity of these shifts. Changes in normal values were also shown to be non-linear, with most of the non-linearity attributed to the shift from growth to maturity. Together, these finding demonstrate that reversed translational bioinformatics may contribute to basic research.
Introduction
Despite the fact that the word ‘translation’ is not inherently directional, Translational Medicine (TM) is normally understood to mean the translation of scientific discoveries made at the bench to improvement in bedside treatment. Reversed TM involves translation in the reverse direction, namely the flow of information and knowledge from the bedside to the bench. Reversed TM is not a new branch of research; at least 57 papers have been published with the words “bedside to bench” included in their title (see for example(1)). In bioinformatics, however, this approach is still emerging.
Given the rapid increase in availability of data generated at bedside, we propose to establish reverse Translational BioInformatics (reversed TBI) as a new field that combines bioinformatics and medical informatics. Bioinformatics offers a toolbox suitable for analyzing large arrays of noisy, poorly annotated and error-ridden biological data (e.g. in transcription profiling(2) or in proteomics(3)). Medicine, on the other hand, brings measurements of a battery of biomarkers for very large populations. In comparison to medicine, even the largest efforts to measure biomarkers in mammals (e.g., the mouse phenome project) involves the measurement of only a few hundred traits in a few hundred animals at one or two ages(4). Mining of clinical literature placed medical informatics at the forefront of knowledge discovery research (5); mining of clinical data may make Homo sapiens an important model system in biology for research areas such as individual variability or aging.
In clinical research, the use of bedside data is becoming ever more common (see for example (6)), although there is an on-going debate on the value of retrospective studies in making clinical decisions(7). But the use of bedside data is not without its challenges. It is conceived as inaccessible and/or of too poor quality, despite an ever increasing number of publications based on bedside data (see for example(8)). Another source of concern is the uncertainty regarding the suitability of existing tools and methodologies for bedside data. Will mining these data necessitate the development of new methodologies and tools, or will existing tools do? Is data quality sufficiently high to justify the effort of accessing and analyzing it?
To partially address these questions, we present a study of the interplay between age, gender and blood biomarkers variation using bioinformatics tools. Clinically, biomarker variation during aging was analyzed in an effort to predict overall disease propensity, with limited success(9). By contrast, the variation in blood biomarkers during growth has not been characterized to the same extent. For example, a recent analysis of the relationship between age and platelet parameters, perhaps the most comprehensive and detailed analysis of this relationship to date, does not include children under 17 (10). In this paper we demonstrate how aging and its interaction with gender can be characterized using clinical data from human. We used the 3rd National Health and Nutrition Examination Survey (NHANES3) (11) as a proxy for bedside data; the study if aging, blood biomarkers, and gender using bioinformatics approaches is used to demonstrate of the reverse TBI approach.
Methods
DATA SOURCE:
The 3rd National Health and Nutrition Examination Survey (NHANES3) was used as the data source, as no source of bedside data is freely and publicly available. NHANES3 was based on a random sample of the non-hospitalized US population comprising 29,314 individuals (13,980 males, 15,334 females). The laboratory section of the NHANES3 public data release was parsed into a tab-delimited text file, and ‘null but meaningful’ values were replaced with null values. The name and abbreviated label of all the biomarkers are given in the legend of Figure 1.
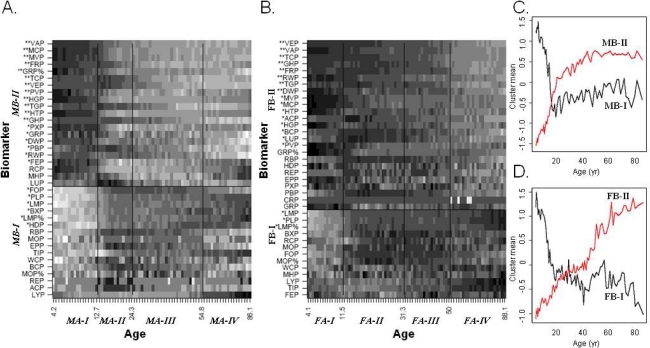
Figure 1.
K-means clustering of age groups and blood biomarkers. Males (A) and females (B) from the NHANES3 survey were divided into equal size groups (N=150). A derived matrix of row-normalized median values for each biomarker in each age group was independently clustered with the K-means algorithm for each age group, assuming 4 such clusters, and biomarkers, assuming 2 such clusters. Cluster borders are indicated by lines and denominated by italics labels on the appropriate axis. The cluster means for men (C) and women (D) in each age group is also shown for the biomarker clusters (red and black lines correspondingly). Biomarkers are sorted within each cluster (from top to bottom) in decreasing covariance with the cluster mean, with 1 or 2 stars indicating biomarkers with r2≥0.2 or r2≥0.4, correspondingly. Abbreviations: ACP, serum alpha carotene; BCP, serum beta carotene; BXP, serum beta cryptoxanthin; CRP, serum C-reactive protein; DWP, platelet distribution width (%); EPP, erythrocyte protoporphyrin; FEP, serum iron; FOP, Serum folate; FRP, serum ferritin; GHP, glycated hemoglobin (%); GRP, granulocyte number (Coulter); GRP%, segment neutrophil (% of 100 cells); HDP, serum HDL cholesterol; HGP, hemoglobin; HTP, hematocrit (%); LMP, lymphocyte number (Coulter); LMP%, lymphocytes (% of 100 cells); LYP, serum lycopene; LUP, serum lutein/zeaxanthin; MCP, mean cell hemoglobin; MHP, mean cell hemoglobin concentration; MOP, mononuclear number (Coulter); MOP%, monocytes (% of 100 cells); MVP, mean cell volume; PBP, serum lead; PLP, platelet count; PVP, mean platelet volume; PXP, serum transferrin saturation; RBP, RBC folate; RCP, red blood cell count; REP, serum sum retinyl esters; RWP, red cell distribution width; TCP, serum cholesterol; TGP, serum triglycerides; TIP, serum TIBC; VAP, serum vitamin A; VEP, serum vitamin E; WCP, white blood cell count.
DATA PROCESSING:
Some fields were eliminated from the original data: (1) irrelevant fields (e.g. ethnicity fields or sample weight fields) or fields duplicated with different units (e.g., PLPSI and PLP), (2) non-numeric or non-continuous fields (i.e. fields with 10 or less different values), (3) fields where more than 1/3 of the values in the raw observation are missing. Of the original 175 fields, 37 and 38 fields remained (for males and females correspondingly) after this elimination, and were used as biomarkers in subsequent analysis. After eliminating individuals missing over 50% of the biomarker measurements, equal size age-gender groups were created including 150 individuals each. Each bin was considered as a single observation, with all the individuals in that bin being considered as biological replicas. The mean age of the individuals in the each age-gender bin was taken as the representative age of the bin. As a result of the binning process, age bins for males and females slightly differed. Hence, for analysis which required comparison by age across the genders, bins of least difference were considered equivalent. For each measurement, a median was calculated, unless measurements were available for less than 5 individuals in a particular age-gender bin (in which case a NULL value was assigned for that biomarker in that bin, with that age bin being removed from subsequence analysis). Each biomarker was than scaled from 0 to 1 for all ages and each gender separately, and Z-transformed for normalization.
K-MEANS CLUSTERING AND MACHINE LEARNING:
The R implementation of the K-means algorithm(12) was used. For the clustering of age groups, the number of means (4) was chosen so as to include one life period over the trivial groups (growth, adulthood, aging). For the biomarkers clustering, the number of means (2) was chosen via trial and error, with attempts to use larger numbers of means yielded clusters that were obviously split in one gender compared other (i.e. one cluster in one gender could be mapped to a pair of clusters in the other). The clementine (SPSS, Chicago, IL USA) implementation of ANN and MLR was used for machine learning.
SUPPLEMENTARY MATERIALS:
A complete description of the algorithms and procedures used in this work are provided in the following website: http://bioinfo.bgu.ac.il/rubin/Misc/rtbi.supp.htm. In this website, scripts are provided (for R and Clementine) that produce all the results from raw NHANES3 data, as well as some of the outputs (i.e. the age specific medians used to generate figures 1 and 2).
Results
The normal values of 37 clinical biomarkers from different age-gender groups were estimated from the NHANES3 survey. The standard-normal scaled medians of age-gender bins were clustered using the K-means algorithm (Figure 1). The basic temporal structure of the data was faithfully reproduced despite the use of a non-temporal clustering algorithm, with consecutive ages clustering together. Age groups that cluster together are more similar (in their biomarker profile) to each other than to other age groups; the border between two clusters may thus point to a change points in human development, in which multiple biomarkers change. The change points indicated from blood biomarkers clustering involve 12.7, 24.3 and 54.8 years for males and 11.5, 30.3 and 50 years for females. In terms of biomarkers clustering, the resulting clusters were generally consistent between males and females. Clusters MB-II and FB-II were highly consistent, with all but 3 biomarkers joined in males also joined in the corresponding female cluster, and with those biomarkers that were relatively highly correlated with their respective cluster means (i.e. r2≥0.4, marked with two stars in Figure 1) in either gender being found both clusters without exception. Clusters MB-I and FB-I were less consistent, with MB-I being bigger than FB-I (17 biomarkers compared to 13). Nevertheless, 10 biomarkers are common to both clusters (58% and 77% of MB-I and FB-I correspondingly), and 100% of the 6 key biomarkers were found in both clusters (i.e., biomarkers with r2≥0.2 with their corresponding cluster mean in either gender; denoted by 1–2 stars in Figure 1).Despite the similarities amongst the clustering of biomarkers by age, gender differences in the overall trend of each cluster were also observed (Figure 1C and 1D), as were differences in the strength of the correlation between each biomarker to its cluster mean (Figure 1A and 1B). In males, cluster MB-I exhibited a sharp decline in ages 4 to 20, which was also observed in FB-I, but the moderate increase, or no change, observed in ages higher than 20 years for cluster MB-I were not observed in cluster FB-I. Cluster MB-II showed a near-linear increase during ages 4–20, which was also observed in cluster FB-II, but the decline in the rate of increase and saturation observed in cluster MB-II starting at 20 years of age, was not observed in cluster FB-II, which presents no clear shift in trend at the age of 20 years. Differences were also in the strength of correlation between each biomarker and its corresponding cluster mean. Several biomarkers with a high coefficient of correlation (r2≥0.4) in one gender only showed moderate (0.2≤r2≤0.4) coefficient of correlation in the other. For example, serum lead levels (PBP) has a higher coefficient of correlation in males than in females (r2 of 0.3 and 0.11 correspondingly; data not shown).
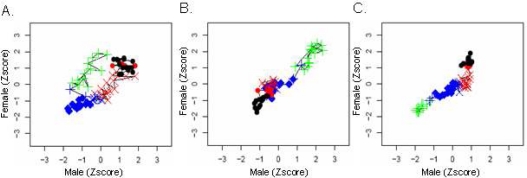
The gender specific patterns of change in biomarkers were further perused using time-course plots that directly compare changes of each biomarker over the lifetime of males and females (Figure 2). In this figure, diagonal lines, such as observed for clusters FA-I and MA-I in serum lead levels (Figure 2A), indicate a concordant trend in both genders. Horizontal or vertical lines, such as observed for serum vitamin A levels (Figure 2B) in clusters FA-III and MA-III, indicate larger variation in one gender relative to the other. If the lines ‘double-back’, this indicates random variation in one group but little variation in the other; if the lines are drawn as ‘dotson-a-string’, they indicate the existence of a trend in one gender, with little or no change in the other. Areas characterized by non-ordered shifts in path direction indicate no trend in either gender, such as observed for platelet counts in all of cluster FA-III and all of cluster MA-III. These three examples suggest that changes in relative biomarkers levels follow a complex and non-linear pattern of change.
Figure 2.
Gender differences in biomarker changes during growth and aging. The age-specific median of serum lead levels (A), platelet counts (B), and serum vitamin A levels (C) is plotted for males (X axis) and females (Y axis). Each value was z-transformed (i.e. subtracting the mean and dividing the difference by the standard deviation of the each biomarker). Each point represents a comparison of similar age bins in the two genders, and is connected with a line to the previous and next age group. The shape of each point represents the cluster it belongs to in males (MA-I, plus; MA-II, diamond; MAfli-III, X; MA-IV, dot), and its color represents females clusters (FA-I, green; FA-II, blue; FA-III, red; FA-IV, black).
This suggestion was further pursued by using machine learning methods to predict age from biomarkers measurements. The nature of the dependency between age and blood biomarkers was gauged by comparing the performance of linear and non-linear. The performance of Artificial Neural Network (ANN), which can model highly non-linear relationships, was compared to Multiple Linear Regression (MLR), a predictors that is best suited for linearly seperable data. The NHANES3 population was thus studied, training ANN and MLR models on 2/3rds of the data and testing on the remaining 1/3. The coefficients of correlation shows that ANN outperformed MLR (Table 1), but only if children and adults are mixed. Comparing the performance of ANN and MLR for adults only, using increasingly homogenous age groups (Table 1) showed that for adults >20 years old, ANN performance was only marginally better than MLR, with the gap becoming even smaller as the age span is restricted to older individuals.
Table 1.
Linear and non-linear prediction of age from biomarkers. Male ages were predicted by training a linear model (MLR) and a non-linear model (ANN) on a training set from the NHANES3 data, and applying them to a test set. The number of individuals (N) and the correlation coefficient (r2) between the predicted and observed ages are given for each classifier.
| Age threshold | N | ANN | MLR |
|---|---|---|---|
| 0 | 4,174 | 0.832 | 0.666 |
| 20 | 2,484 | 0.594 | 0.504 |
| 40 | 1,514 | 0.437 | 0.353 |
Discussion
The relationship between age, gender and blood biomarkers was studied utilizing data from the NHANES3 survey. Adopting a quick and naïve bioinformatics approach to the analysis allowed us to make many observations regarding human growth and aging which can later be checked with more rigorous methodologies. Using K-means clustering to cluster age groups according to their similarity in median biomarker levels, some points in the human life-course were shown to be reflected in the blood biomarker profile (Figure 1). Some of the change points found by clustering may correspond to well characterized physiological processes (e.g. ages 11.5 and 12.7 for males and females corresponding to the peak of growth at puberty, occurring at ages 11.5 and 13.5 correspondingly(13); age 50 in females corresponding to menopause, with a median at 50–51(14); age 24.3 in males corresponding to growth completion at 25). The remaining critical ages (31.3 in females and 54.8 in men) may represent novel critical ages, or an artifact of the K-means clustering algorithm. Clustering of biomarkers across the ages, on the other hand, defines a grouping of blood biomarkers, involving trivial biomarkers combinations (such as serum level of beta cryptoxanthin and lycopene), but also including some intriguing relations, such as the clustering of lead with specific vitamins (vitamins A and E). Such observations can potentially impact many research areas. For example, it may encourage specific models of age-dependent changes in population serum lead levels(15).
The direct comparison of life-course changes in individual biomarker between males and females was demonstrated as a potential tool for uncovering gender-specific trends (Figure 2). This analysis suggested the biomarkers interact non-linearly with age; this observation could be further persued thanks to the large sample size NHANES3 provides. Comparing linear and non-linear models using increasing age thresholds showed that the nonlinearity of biomarker-age interactions can largely be attributed to the shift from growth to aging (Table 1).
The analysis presented here is naïve in many ways. K-means clustering is hardly the most sensitive or robust clustering algorithm. For the biomarkers clustering, for example, temporal algorithms (e.g. (16)) are likely to provide more sensitive and reliable results. Moreover, proper statistical scrutiny still needs to be applied the findings before they are to be further considered. Nevertheless, this naïve analysis demonstrates how the bioinformatics tools can be directly applied to clinical data, and that clear trends can be identified in clinical data without the tight control that only prospective studies or model animals provide.
How suitable is bedside data for the type of analysis presented here? A debate about its usefulness for other applications, such as laboratory reference values calculation(17), provides useful lessons. At the heart of the debate is the potential impact of outlier patients on estimated 95% population intervals, information that is critical to the care giver but not the basic biologist. As demonstrated here, biological trends can be identified using the median, which is much more robust to outliers; it can thus be expected that if bedside data is even marginally successful in determining 95% intervals, bedside data can be very useful for the less demanding job of trend and pattern analysis. Bedside data also offers its own advantages over public surveys, such as involving huge populations with possibly better coverage of older ages. In addition, bedside data contains multiple measurements for the same individual, which can greatly improve variation estimates, and as a result may allow better quantitative models to be developed. To conclude, we demonstrate how classical bioinformatics approaches can serve to exploit clinical data for the study of human biology. As such, we show that reverse-TBI can contribute to basic research by opening up new and unique sources of data.
Acknowledgments
We wish to thank Dr. Vadim Fraifeld from Ben Gurion University for fruitful discussions. This work was supported by the Horowitz Center for Complexity Science, the RICH foundation and the National Institute of Biotechnology in the Negev.
References
- 1.Wessler S, Gitel SN, Salzman E, Deykin D, Licht J, Freedberg AS, et al. Warfarin - From Bedside To Bench. New England Journal of Medicine. [Discussion] 1984;311(10):645–52. doi: 10.1056/NEJM198409063111007. [DOI] [PubMed] [Google Scholar]
- 2.Quackenbush J.Microarray data normalization and transformation Nat Genet 2002. 2002 December32Suppl496–501. [DOI] [PubMed] [Google Scholar]
- 3.Listgarten J, Emili A.Statistical and computational methods for comparative proteomic profiling using liquid chromatography-tandem mass spectrometry Mol Cell Proteomics 2005. 2005April44419–34. [DOI] [PubMed] [Google Scholar]
- 4.Bogue M, Grubb S.The Mouse Phenome Project Genetica 2004. 2004 September122171–4. [DOI] [PubMed] [Google Scholar]
- 5.Swanson DR.Fish oil, Raynaud’s syndrome, and undiscovered public knowledge Perspect Biol Med 1986. 1986 3017–18. [DOI] [PubMed] [Google Scholar]
- 6.Chen D, Weber S, Constantinou P, Ferris T, Lowe H, Butte A. Novel integration of hospital electronic medical records and gene expression measurements to identify genetic markers of maturation. Pac Symp Biocomput. 2008:243–54. [PMC free article] [PubMed] [Google Scholar]
- 7.Ioannidis J, Haidich A, Pappa M, Pantazis N, Kokori S, Tektonidou M, et al. Comparison of evidence of treatment effects in randomized and nonrandomized studies JAMA 2001. 2001 August2867821–30. [DOI] [PubMed] [Google Scholar]
- 8.Tirosh A, Shai I, Tekes-Manova D, Israeli E, Pereg D, Shochat T, et al. Normal fasting plasma glucose levels and type 2 diabetes in young men. New England Journal of Medicine. [Article] 2005 Oct;353(14):1454–62. doi: 10.1056/NEJMoa050080. [DOI] [PubMed] [Google Scholar]
- 9.Johnson TE. Recent results: Biomarkers of aging. Experimental Gerontology. [Review] 2006 Dec;41(12):1243–6. doi: 10.1016/j.exger.2006.09.006. [DOI] [PubMed] [Google Scholar]
- 10.Segal J, Moliterno A.Platelet counts differ by sex, ethnicity, and age in the United States Ann Epidemiol 2006. 2006 February162123–30. [DOI] [PubMed] [Google Scholar]
- 11.USA DoHaHSD Plan and operation of the Third National Health and Nutrition Examination Survey, 1988–94. Series 1: programs and collection procedures Vital Health Stat 1 1994. 1994 July321–407. [PubMed] [Google Scholar]
- 12.Hartigan J, Wong M. Algorithm AS 136: A K-Means Clustering Algorithm. Applied Statistics. 1979;28(1):100–8. [Google Scholar]
- 13.Abbassi V. Growth and normal puberty. Pediatrics. [Article] 1998 Aug;102(2):507–11. [PubMed] [Google Scholar]
- 14.Braunwald E, Fauci A, Kasper D, Hauser S, Longo D, Jameson J. Harrison’s principles of internal medicine. New York, NY: McGraw Hill; 2001. [Google Scholar]
- 15.Bressler JP, Olivi L, Cheong JH, Kim Y, Maerten A, Bannon D. Metal transporters in intestine and brain: their involvement in metal-associated neurotoxicities. Human & Experimental Toxicology. [Article] 2007 Mar;26(3):221–9. doi: 10.1177/0960327107070573. [DOI] [PubMed] [Google Scholar]
- 16.Ma P, Castillo-Davis C, Zhong W, Liu J. A data-driven clustering method for time course gene expression data. Nucleic Acids Res. 2006;34(4):1261–9. doi: 10.1093/nar/gkl013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Horn PS, Pesce AJ. Reference intervals: an update. Clinica Chimica Acta. [Review] 2003 Aug;334(1–2):5–23. doi: 10.1016/s0009-8981(03)00133-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
A complete description of the algorithms and procedures used in this work are provided in the following website: http://bioinfo.bgu.ac.il/rubin/Misc/rtbi.supp.htm. In this website, scripts are provided (for R and Clementine) that produce all the results from raw NHANES3 data, as well as some of the outputs (i.e. the age specific medians used to generate figures 1 and 2).