Abstract
Chronic Lymphocytic Leukemia (CLL) is the most common adult leukemia in the U.S., and is currently incurable. Though a small number of biomarkers that may correlate to risk of disease progression or treatment outcome in CLL have been discovered, few have been validated in prospective studies or adopted in clinical practice. In order to address this gap in knowledge, it is desirable to discover and test hypotheses that are concerned with translational biomarker-to-phenotype correlations. We report upon a study in which commonly available ontologies were utilized to support the discovery of such translational correlations. We have specifically applied a technique known as constructive induction to reason over the contents of a research data repository utilized by the NCI-funded CLL Research Consortium. Our findings indicate that such an approach can produce semantically meaningful results that can inform hypotheses about higher-level relationships between the types of data contained in such a repository.
Introduction
Chronic Lymphocytic Leukemia (CLL) is the most common adult leukemia in the United States, and is associated with an increasing incidence rate [1]. Due to its highly heterogeneous clinical course and phenotypic presentation, there are no known curative strategies. As such, current clinical best practices emphasize delaying treatment until a patient demonstrates either symptomatic or progressive disease, an approach that does not necessarily correlate with optimal treatment outcomes or long-term survival [2]. Recent studies have identified several bio-molecular markers, including leukemic-cell expression of CD38 surface markers, mutational status of immunoglobulin heavy chain variable region genes (IgVH), zeta-chain associated protein (ZAP-70) expression level and specific chromosomal abnormalities, which can be used to identify those patients most at risk or requiring early intervention for progressive CLL [2]. While these bio-molecular markers have been demonstrated in a small number of studies to significantly correlate with risk of progressive disease, none have been shown to correlate with treatment outcome in prospective clinical trials [2]. Given the benefits of employing adaptive therapies based upon a patient’s phenotypic characteristics as established with other hematologic malignancies, the further exploration and validation of prognostic bio-molecular markers in CLL is highly desirable.
The utility of informatics-based approaches to the discovery and validation of biomarker-to-phenotype correlations has been reported in the literature on numerous occasions. One such approach involves the use of conceptual knowledge engineering techniques to identify potential relationships between data types in large-scale, multidimensional biomedical data sets [3]. Conceptual knowledge engineering targets the identification and manipulation of conceptual knowledge structures or collections that consist of atomic units of knowledge, or “facts” (e.g., “increased white blood cell count”, “chromosome 11 abnormality”, etc.) and the network of relationships among those units [4].
We report upon the application of such conceptual knowledge engineering techniques in order to address the gap in knowledge concerning the identification of biomarkers capable of supporting adaptive therapy in CLL. Given this motivation, in the following sections we will:
Provide additional definitions and details on conceptual knowledge engineering-based approaches to knowledge discovery in databases,
Introduce an experimental context for our work, specifically a collaboration with the CLL Research Consortium (CRC), and
Present results from a feasibility and validity evaluation in which a knowledge discovery in databases approach known as constructive induction was used to support the analysis of a CLL-specific multi-dimensional bio-molecular and phenotypic data set.
The specific aim of the feasibility and validity evaluation that we will report on is to develop a conceptual knowledge collection that corresponds to the contents of a research data repository maintained by the CRC, and in doing so evaluate the hypothesis that constructive induction is a feasible technique for generating valid and potentially novel hypotheses concerning bio-marker-to-phenotype relationships that are comprised of “facts” corresponding to the contents of the CRC research data repository.
Background
The following section further describes the specific conceptual knowledge engineering techniques used in our study, as well as the experimental content:
Knowledge Engineering (KE) is a process by which knowledge is collected, represented and subsequently used by computational agents to replicate expert human performance in an application domain. Conceptual knowledge, one of three primary types of knowledge that can be targeted by KE, can be defined as a combination of atomic units of information (e.g., “facts”) and the meaningful relationships among those units [4]. Conceptual knowledge collections in the biomedical domain include ontologies, controlled terminologies, semantic networks and database schemas.
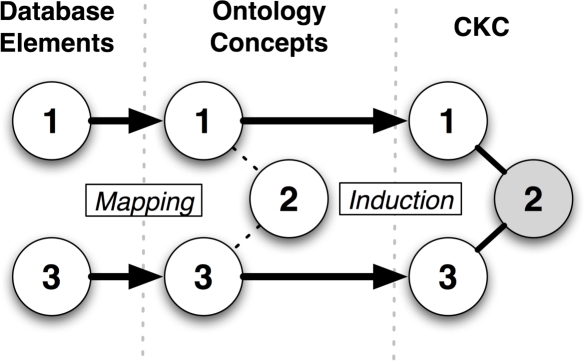
Knowledge discovery in databases (KDD) is a specific type of conceptual knowledge engineering technique that is used to elicit both atomic units of knowledge and the relationships among them from the contents of a database construct [4]. Domain-specific knowledge collections, such as ontologies, are commonly used during KDD in order to augment meta-data contained in the targeted database schema. This overall approach is the basis for a specific KDD methodology known as constructive induction [5] (Figure 1).
Figure 1:
Constructive induction methodology.
In constructive induction, distinct data types (i.e., “facts”) defined by a database schema are mapped to concepts defined in one or more ontologies. Subsequently, the relationships included in these ontologies are used to induce semantically meaningful linkages between the mapped data types. If a concept is included in the ontology, but does not map to a data type in the database, it can be used as an intermediate concept in order to induce new semantically related concept triplets or high-order relationships that begin and terminate with data types contained in the source database schema. By exploiting the transitive closure principle that is applicable when representing an ontology as a graph system, this induction process generates conceptual knowledge constructs (CKCs) that are defined in terms of data elements and the semantic relationships that link them together, as encoded in the source ontologies being utilized. The resulting CKCs can then be used to inform potential hypotheses about relationships between data types contained in the source database [5].
The CLL Research Consortium (CRC; http://cll.ucsd.edu) is an NCI-funded research consortium consisting of eight sites, which coordinate and facilitate basic and clinical research on the genetic, biochemical and immunologic bases of CLL. The ultimate goal of the CRC is to discover and evaluate novel biologic and pharmacologic treatments for CLL, and examine phenotypic ↔ bio-molecular relationships that may improve clinical staging and/or assist in evaluating patient responses to such novel therapies. The CRC utilizes an integrated information management system, known as CIMS, that incorporates a shared data repository and multiple task-specific web portal interfaces supporting clinical trials, basic science and tissue bank data management. Currently, CIMS is being used to collect, manage and analyze data for over 4000 patients involved in multiple clinical trial modalities, as well as hundreds of thousands of CLL-specific tissue samples. Given its unique capabilities, the CRC is well positioned to investigate high-priority bio-molecular markers (e.g., ZAP70 expression levels) utilizing large-scale basic science, clinical and epidemiologic studies. However, doing so relies heavily on the ability to reason about and infer meaning from the data collected via CIMS.
Methods
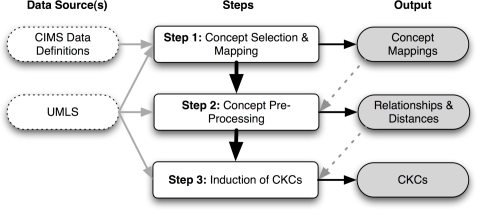
The following sections summarize a three-step methodology (Figure 2) that was employed and formal validity evaluation that was conducted during the course of the study reported on in this manuscript.
Figure 2:
Overview of study methods
Step 1 - Concept Selection and Mapping
A corpus of phenotypic and bio-molecular data element definitions was extracted from the CIMS data repository model. A subject matter expert (SME) used the Unified Medical Language System Knowledge Source Server (UMLSKS) [6] free text search engine and The Systematized Nomenclature of Medicine Clinical Terms (SNOMED-CT) [7] CliniClue browser [8] to map each data element to one or more UMLS concepts corresponding to the SNOMED-CT [7] and NCI Thesaurus [9] source vocabularies. The mapped concepts were heuristically selected such that they either described the action resulting in the data element (e.g., laboratory procedure such as white blood cell count); and/or the specific values that could be contained within a particular database field (e.g., laboratory test results such as a value indicating an increased white blood cell count).
Step 2 - Concept Pre-processing
The UMLS MRHIER source file enumerates all unique hierarchical paths (determined by the source vocabulary) between a given UMLS concept and the UMLS root concept (represented as an atomic string). If the source vocabulary allows for multi-hierarchies, a concept may have more than one path to the root. Using this file, the minimum distance (i.e., number of “steps” or atoms) to the root was calculated for each UMLS concept corresponding to either the SNOMED-CT or NCI Thesaurus source vocabularies. This information is used in Step 3 to facilitate the application of a variable search depth parameter (d), which is used as a surrogate indicator of concept granularity. In addition, a subset of the UMLS MRREL source file, which contains information regarding all possible relationships between two given UMLS concepts, was created by: 1) extracting only parent, child and semantic relationships between concepts corresponding to the SNOMED-CT or NCI Thesaurus source vocabularies 2) determining inverse relationships within the previously selected subset (e.g., diagnoses is the inverse of diagnosed_by); 3) further refining the subset of selected relationships to encompass only those thought to be most meaningful for relating bio-molecular and phenotypic concepts as judged by two SMEs.
Step 3 - Induction of CKCs
The relationship table generated in Step 2 was employed to determine any pair-wise semantic relationships between concepts that corresponded to mapped data types in the CIMS data repository (e.g., del(11q22q23) - may be cytogenetic abnormality of disease - chronic lymphocytic leukemia refractory). These pair-wise relationships were then transitively expanded to generate CKCs that included up to three intermediate concepts. Any CKC where the rightmost, or terminating concept corresponded to a CRC repository data type was recorded for later analysis. With the addition of each transitively related concept, a validation algorithm employing an index of inverse relationships (as defined in Step 2) was used to ensure that subsequent relationships were not inverses of each other, and no duplicate concepts or cycles occurred within the resulting CKCs. To explore varying levels of granularity in the induced CKCs, all possible transitive relationships between concepts were iteratively calculated with d (i.e., minimum distance from the root as calculated in Step 2) set at values from 1–6, where each concept in the CKC must have a distance from the root ≥ d.
Validity Evaluation
An evaluation of the results generated in the preceding steps was conducted to assess the 1) accuracy of the concept codes manually assigned to the CIMS data elements, and 2) validity and meaningfulness (i.e., could the concepts and relationships be used to inform a hypothesis) of the induced CKCs. For the first phase of the evaluation, individuals with expertise in database design and biomedical terminologies were asked to evaluate a randomly selected set of CIMS database elements and the ontology concepts to which they were mapped with respect to the accuracy of those mappings given the heuristics employed in Step 1. During the second phase of the evaluation, a d x n matrix evaluation was constructed, where a single CKC from each search depth (d) and number of included concepts (n) was randomly selected. CLL SMEs (basic science or clinical investigators) were asked to assess each CKC for both validity (using a categorical response of: Completely Valid, Partially Valid/Invalid, or Completely Invalid) and meaningfulness (using a categorical response of Meaningful or Not Meaningful).
Results
Step 1 - Concept Selection and Mapping
A corpus of 107 data elements was extracted from the CIMS data repository schema, of which 68 (63.5%) and 39 (36.4%) corresponded to phenotypic and bio-molecular parameters, respectively. These data elements mapped to a total of 882 UMLS concepts (537 unique), of which 455 (51.6%) corresponded to the initial phenotypic parameters and 427 (48.4%) corresponded to the initial bio-molecular data elements. Examples of the phenotypic concepts selected during this process included white blood cell count and disease-specific performance status, while examples of the bio-molecular concepts included leukemic cell CD5 frequency and chromosome 11 abnormality.
Step 2 - Concept Preprocessing
The average distances from the root for the previously selected concepts corresponding to phenotypic and bio-molecular parameters were found to be 5.5 [range: 3–10] and 5.5 [range: 2–10], respectively. Using the methods described earlier, a total of 196 unique UMLS semantic relationships (e.g., ‘may be cytogenetic abnormality of disease’, ‘disease may have abnormal cell’, ‘has definitional manifestation’, ‘disease has finding’) were selected for subsequent use.
Step 3 - Induction of CKCs
During this final step, CKCs with 2 ≤ n ≤ 5, where n is the number of concepts contained within a CKC, linking the bio-molecular and phenotypic concepts selected in Step 1, were induced iteratively at increasing values of d, as summarized in Table 1.
Table 1:
Summary of number of CKCs stratified by search depth (d) and the number of concepts (n).
| Ontology Search Depth | Number of Concepts inCKCs | ||||
|---|---|---|---|---|---|
| 2 | 3 | 4 | 5 | Total | |
| 1 | 5 | 896 | 844 | 139,024 | 140,769 |
| 2 | 5 | 676 | 822 | 136,456 | 137,959 |
| 3 | 5 | 676 | 822 | 136,456 | 137,959 |
| 4 | 5 | 676 | 804 | 133,816 | 135,601 |
| 5 | 5 | 145 | 351 | 8,656 | 9,157 |
| 6 | 0 | 3 | 57 | 3,063 | 3,063 |
A complete set of the selected concepts, their distance from the ontology root, and semantic relationships for all search depths is available at http://bmi.osu.edu/~payne/crcokd.html.
Validity Evaluation
Five SMEs evaluated the validity of the mappings between data types and ontology concepts. The evaluation sets were non-overlapping (i.e., a total of 250 unique mappings were evaluated). The SMEs completely agreed with 173 (69.2%), partially agreed/disagreed with 40 (16%) and completely disagreed with 6 (2.4%) of these concept mappings. The SMEs felt that they did not have the domain expertise or enough information to assess a total of 31 (12.4%) of the selected concept mappings.
Five SMEs evaluated the validity and meaningfulness of the induced CKCs. Each SME was given a total of 23 CKCs to evaluate, due to the fact that there were no pair-wise relationships generated at d = 6. A total of 49 (43%) of the 115 selected CKCs were designated as non-evaluable by the SMEs due to the fact that they did not have sufficient domain expertise to adequately assess a given association, particularly with respect to the intermediate concepts. Of the remaining 66 CKCs, the SMEs concluded that 16 (24.2%), 43 (65.2%) and 7 (10.6%) were completely valid, partially valid/invalid and completely invalid, respectively. A qualitative review of the SME’s comments indicates that in most cases where a CKC was designated partially valid/invalid, the experts felt that potentially informative intermediate concepts had been omitted from the construct. Finally, 10 of the completely valid CKCs were assessed for meaningfulness (one SME omitted this section of the survey), and of these, 9 (90%) were deemed to be meaningful (Table 2). The average distance from the ontology root for concepts included in such CKCs was found to be 7 (SD=1).
Table 2:
Examples of valid/ meaningful CKCs
| Relationship Pattern | Induced Relationship |
|---|---|
| Chromosomal Abnormality → Diagnosis | del(17p13) – [may be cytogenetic abnormality of disease] - Chronic lymphocytic leukaemia refractory |
| Chromosomal Abnormality → Clinical Laboratory Value/Finding | t(6;9)(p23;q34) - [may be cytogenetic abnormality of disease] - Acute Myelomonocytic Leukemia without Abnormal Eosinophils- [disease may have finding] - White blood cell count increased |
Discussion
We have described the application of a type of conceptual knowledge engineering technique known as constructive induction in order to generate conceptual knowledge constructs (CKCs) comprised of phenotypic and bio-molecular concepts that correspond to the contents of the CIMS data repository. The objective of this induction process is to support higher-order reasoning about the contents of the CIMS repository, with the specific goal of discovering potentially informative biomarker-to-phenotype relationships. Our results lead to several findings, specifically:
Constructive induction is computationally tractable for generating biomarker-to-phenotype CKCs derived from ontology-anchored concepts that correspond to source data elements in the CIMS repository and semantic inter-relationships.
The semi-automated mapping of data elements to ontology concepts was generally accurate, with SMEs at least partially agreeing with such associations 85.2% of the time. However, there is clear room for improvement in this step of the methodology.
In 89.4% of evaluable instances, the induced CKCs were at least partially valid, with 24.2% of the CKCs being designated completely valid. A qualitative review of the SME’s comments indicates that those CKCs designated as partially valid contained valid concepts and relationships, but were not considered complete (e.g., they lacked important intermediate concepts or relationships per the experts’ judgment). Such an outcome would appear to point to potential shortcomings of the source ontologies. Despite this possible limitation, when CKCs were found to be completely valid, they were usually meaningful (90% of the time), indicating that they could be useful in informing hypotheses concerning translational biomarker-to-phenotype relationships. Of interest, the concepts that comprised CKCs designated as completely valid and meaningful were found to have a depth from the ontology root equal to or greater than the depths of the initial and terminating concepts used to induce those constructs. This suggests a possible correspondence between search depth constraints and meaningfulness that could be used to identify those CKCs more likely to aid in the discovery of informative biomarker-to-phenotype relationships.
There are several limitations to the work reported in this manuscript, including the: 1) use of a semiautomated and difficult to scale human-mediated process for mapping database elements to ontology concepts; 2) use of a surrogate indicator of concept granularity within source terminologies derived from distance-to-root metrics; 3) use of relatively simple graph-theoretic reasoning techniques to induce CKCs, and 4) limits on our evaluation study imposed by the domain expertise of the recruited SMEs In order to address the first of these potential limitations, we are assessing techniques for automating the database element-ontology mapping process using tools such as MetaMap (mmtx.nlm.nih.gov) and programmatic interfaces to the UMLSKS Server. Similarly, to address the second and third of the preceding limitations, we are actively evaluating W3C Semantic Web [10] initiative-derived platforms, including: 1) OWL-based ontology representations; and 2) the Jena semantic web framework, in order to apply advanced ontology reasoners to our CLL-specific data set. Unfortunately, due to a paucity of ontologies represented using OWL, which also exhibit sufficient content coverage of the data types used in this study, additional work will be required to transform non-OWL compliant ontologies into compatible formats in order to support such an approach.
Conclusion
The preceding results serve to demonstrate that conceptual knowledge engineering techniques, and in particular constructive induction, can reasonably be applied to the contents of an operational translational research data repository. This approach can support higher-order reasoning about relationships between data elements in such a repository, based upon knowledge encoded in commonly available biomedical ontologies. The results of our validity evaluation indicate that in those cases where the content coverage of the selected source ontologies is sufficient, thus allowing for the induction of valid and complete CKCs, such constructs can be informative to the discovery of biomarker-to-phenotype relationships. However, our results also indicate that there is significant room for improvement in the described methodology in order to: 1) increase the scalability of the initial database element to ontology concept mapping process; and 2) identify and censor potentially less-useful CKCs. Given our findings, we believe that the use of constructive induction to discover potentially informative biomarker-to-phenotype relationships that correspond to the contents of operational data repositories holds great promise. However, it is also clear that additional work is required to refine and optimize such approaches.
Acknowledgments
Support provided by NCI 2 P01CA081534-07A1
References
- 1.Kipps TJ. Immunobiology of chronic lymphocytic leukemia. Curr Opin Hematol. 2003;10(4):312–8. doi: 10.1097/00062752-200307000-00010. [DOI] [PubMed] [Google Scholar]
- 2.Grever MR, et al. Comprehensive assessment of genetic and molecular features predicting outcome in patients with chronic lymphocytic leukemia: results from the US Intergroup Phase III Trial E2997. J Clin Oncol. 2007;25(7):799–804. doi: 10.1200/JCO.2006.08.3089. [DOI] [PubMed] [Google Scholar]
- 3.Sung NS, et al. Central challenges facing the national clinical research enterprise. Jama. 2003;289(10):1278–87. doi: 10.1001/jama.289.10.1278. [DOI] [PubMed] [Google Scholar]
- 4.Payne PR, et al. Conceptual knowledge acquisition in biomedicine: A methodological review. J Biomed Inform. 2007;40(5):582–602. doi: 10.1016/j.jbi.2007.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Joseph P, Bruce GB. Ontology-guided knowledge discovery in databases, in Proceedings of the international conference on Knowledge capture. ACM Press; Victoria, British Columbia, Canada: 2001. [Google Scholar]
- 6.UMLS Knowledge Source Server (umlsks.nlm.nih.gov) 2008. National Library of Medicine. [Google Scholar]
- 7.SNOMED-CT ( www.snomed.org) 2008. College of American Pathologists
- 8.CliniCue ( www.clinicue.com) 2008. CliniCue.
- 9.NCI Thesaurus (ncicb.nci.nih.gov). 2008. National Cancer Institute [Google Scholar]
- 10.W3C Semantic Web ( www.w3.org/2001/sw) 2008. W3C.